Abstract
Traditional regression-based approaches do not provide good results in diagnosis and prediction of occurrences of cardiovascular diseases (CVD). Therefore, the goal of this paper is to propose a deep learning–based prediction model of occurrence of major adverse cardiac events (MACE) during the 1, 6, 12 month follow-up after hospital admission in acute myocardial infarction (AMI) patients using knowledge mining. We used the Korea Acute Myocardial Infarction Registry (KAMIR) dataset, a cardiovascular disease database registered in 52 hospitals in Korea between 1 January, 2005, and 31 December, 2008. Among 14,885 AMI patients, 10,813 subjects in age from 20 to 100 years with the 1-year follow-up traceability without coding errors were finally selected. For our experiment, the training/validation/test dataset split is 60/20/20 by random sampling without replacement. The preliminary deep learning model was first built by applying training and validation datasets and then a new preliminary deep learning model was generated using the best hyperparameters obtained from random hyperparameter grid search. Lastly, the preliminary prediction model of MACE occurrences in AMI patients is evaluated by test dataset. Compared with conventional regression-based models, the performances of machine/deep learning–based prediction models of the MACE occurrence in patients with AMI, including deep neural network (DNN), gradient boosting machine (GBM), and generalized linear model (GLM), are also evaluated through a matrix with sensitivity, specificity, overall accuracy, and the area under the ROC curve (AUC). The prediction results of the MACE occurrence during the 1, 6, and 12-month follow-up in AMI patients were the AUC of DNN (1 M 0.97, 6 M 0.94, 12 M 0.96), GBM (0.96, 0.95, 0.96), and GLM (0.76, 0.67, 0.72) in machine learning–based models as well as GRACE (0.75, 0.72, 0.76) in regression model. Compared with previous models, our deep learning–based prediction models significantly had the accuracy of 95% or higher and outperformed all machine learning and regression-based prediction models. This paper was the first trial of deep learning–based prediction model of the MACE occurrence in AMI clinical data. We found that the proposed prediction model applied different risk factors except the attribute “age” by using knowledge mining and directly used the raw data as input.
Similar content being viewed by others
Explore related subjects
Find the latest articles, discoveries, and news in related topics.Avoid common mistakes on your manuscript.
1 Introduction
According to the World Health Organization (WHO), cardiovascular disease (CVD) was the primary cause of deaths in the world [1]. In 2015, 17.7 million people died of CVD, and it held 31% of deaths in the world. With the publication of the Framingham Heart Study in the 1960s [2], the concept of risk factors for CVD was established and CVD risk prediction models using the risk factors have been studied. So far, data mining techniques of prediction models on CVD occurrences in AMI patients can be cataloged into two different approaches such as regression-based and machine learning–based methods. Currently, Framingham risk score (FRS) [3,4,5], QRISK [6, 7], TIMI [8, 9], and GRACE [10,11,12] models are available in regression-based cardiovascular risk prediction models. Their developments aim to apply for clinical diagnosis by converting the risk factors into risk indices by using the thorough study and personal computation of dataset.
There are many technological features to promote physical activities using smartphone apps [13].
Machine learning techniques are being effectively used in order to collect medical data from divers’ sources [14]. There were also various machine learning–based classification methods for the prediction model of CVD occurrences, such as random forests (RF) [15], neural networks (NN) [16,17,18], and support vector machines (SVM) [19, 20]. Unstructured data of CVD patients was used to identify early heart failure using the Naïve Bayesian machine learning approach [21]. The machine learning–based approaches were known as methods to solve the limitations of traditional regression-based prediction models. The main characteristics of the machine learning–based models were available for finding the associations between different diseases and had the high accuracy of prediction and excellent ability to process missing and outlier datasets using data mining tools and techniques. It was also possible to perform large-scale data analysis by small and incomplete training datasets and many dependent variables which were the disadvantage of the regression-based model (logistic regression and Cox-proportional hazard regression models) [22].
Nevertheless, we can summarize the challenging issues in previous prediction models of CVD occurrence in AMI patients as follows. First, most of the previous regression-based CVD prediction models do not provide good results in the prognosis and diagnosis of the CVD occurrences in patients with moderate risk. For example, approximately half of myocardial infarction (MIs) and strokes will occur in people who are not predicted to be at the risk of CVD [23]. Moreover, previous guidelines for CVD risk showed low prediction accuracy, and thus unnecessary treatments sometimes occurred in patients with moderate risk due to inefficient knowledge of prediction models. Second, we implicitly assumed that each risk factor in regression-based CVD prediction model was related to the occurrence of MACE in AMI patients and the non-linear interactive relations among risk factors were oversimplified [22]. Third, conventional regression-based CVD prediction tools included the major risk factors such as age, blood pressure, heart rate, diabetes, cholesterol, smoking, and history of heart disease, whereas machine/deep learning approaches appeared different risk factors [24]. Fourth, machine/deep learning–based studies have done in various medical areas but mainly focused on analyzing medical images using the convolutional neural network (CNN) [25]. Especially, there was no deep learning–based prediction model for the prognosis and diagnosis of the MACE occurrences on clinical data in AMI patients and there was insufficient knowledge of prediction models in previous research works.
Therefore, this paper proposes a deep learning–based prediction model of the MACE occurrences during the 1, 6, and 12-month follow-up in the AMI clinical data with the aim to develop a knowledge mining–based clinical decision support system and assess the risk of after hospital discharge patients. The detailed research contents of this paper can be summarized as follows. First, our deep learning–based prediction model of the MACE occurrences is generated by applying training dataset and hyperparameter values obtained from the random hyperparameter grid search method on the dataset. Then we obtain many preliminary prediction models that consist of 20 input neurons, three hidden layers, and one output neuron. Second, the preliminary prediction models are evaluated through validation dataset, and then the best prediction model is selected. Third, the selected prediction model is finally evaluated with test dataset. Lastly, the accuracy of proposed prediction model will be compared with GBM, GLM, and GRACE during the 1, 6, and 12-month follow-up after discharge.
2 Method and materials
2.1 Data preparation
The Korea Acute Myocardial Infarction Registry (KAMIR) was the first nationwide, multicenter online registry designed to describe the characteristics and clinical outcomes of patients presenting with myocardial infarction (MI) and reflected current management of patients with acute myocardial infarction (AMI) in Korea [26]. The registry includes 52 community and university hospitals with the capability of primary PCI. Data were collected at each site by trained study coordinators with the standardized protocol retrospectively. For the experiment, this paper used 14,885 AMI subjects enrolled in KAMIR from 1 October, 2005, to 28 February, 2008. Among them, we selected 10,813 patients aged between 20 and 100 years old with the 1-year follow-up MACE after hospital discharge. Excepted the records of 4072 with missing values and with no attribute “MACE” value in the population during the follow-up period, the experimental dataset consisted of 10,813 records × 49 columns with 21 numerical, 26 categorical, and 4 discrete data. The dataset was preprocessed by creating new variables (age, survival time) and reevaluating variables (Killip class, LVF, etc.) before creating the prediction model of MACE occurrences.
2.2 Applied risk factors
The KAMIR data schema has 51 variables as shown in Table 1, and their variables and types are divided into two domains of demographic characteristics and clinical findings. The demographic characteristics are age, gender, height, weight, blood pressure, heart rate, Killip class, heart rhythm, hypertension, diabetes mellitus, pain, dyslipidemia, smoking history, family history of heart disease, history of ischemic heart disease, comorbidities, final diagnosis, and MACE. Clinical features include glucose, creatinine, creatine kinase (CK), CK-MB, Troponin-I, maximum Troponin-I, maximum Troponin-T, total cholesterol, triglyceride, HDL-cholesterol, LDL-cholesterol (hs-CRP), N-terminal brain natriuretic peptide (NT-proBNP), and glycated hemoglobin. The primary endpoints in this study are defined as major adverse cardiac events (MACE) that occurred during the 1, 6, and 12-month follow-up after discharge. We determined the MACE as cardiac death, non-cardiac death, re-PCI, and CABG, and calculated as a patient experienced the MACE not the total of events but at least one or more during the 1, 6, and 12-month follow-up after discharge.
2.3 Training, validation, and test datasets
In case a prediction model either increases the number of parameters or uses a non-linear model such as a neural network model, the prediction performance by training data can be high in a moment due to overfitting. With the overfitting occurrence, the prediction accuracy by training data is high, but the prediction performance by test data can be rapidly reduced. Therefore, to objectively measure the performance of prediction model, our proposed prediction model was evaluated by a new dataset that did not use for training, namely validation and test datasets. Experimental datasets are divided into training dataset of 60% for model learning, validation dataset 20% for model selection, and test dataset 20% for evaluating the final selected model by random sampling without replacement on the bases of knowledge-based observations of dataset.
2.4 Applied machine learning algorithms
For the implementation of prediction model of MACE occurrences in AMI patients, this paper applied three machine learning algorithms: deep neural networks (DNN) [27], GBM [28], and GLM [29]. First, DNN is an artificial neural network (ANN) with multiple hidden layers between the input and output layers, consisting of three hidden layers in artificial networks and non-linear patterns in unstructured data. In this paper, we use the deep feedforward networks which are a quintessential deep learning model of the DNN model. ANN develops the relationship between input layer and output layer using the same architecture as derived from the human brain. All layers comprise of numerous neurons or nodes. The ANN applies a backpropagation algorithm for training process. Second, GBM is the method of boosting method plus gradient descent. It creates a model, generates a fitting model to the residual, and combines both models. Then, in case the residual again finds in the coupled model, then the fitting model creates in the residual, and the final prediction model generates by repeating until the residual does not exist. GBM provides a competitive approach for both regression and classification, especially for classifying less clean data. A forward stage additive model is designed by gradient descent function space to build a regression tree for different features in a distributed way [30]. We performed 50 bosting stages for GBM training. The deviance-loss function has been applied for optimization and Friedman mean-square-error function has been applied measuring splitting quality. Third, GLM is an extension of the linear regression model which enhances the linear model so that it analyzed even when dependent variables are not in the normal distribution. GLM is a combination of traditional statistical methods and machine learning techniques in which dependent variables are linearly related to independent variables through a specified link function and finds the combination of hyperparameter values via the grid search approach. GLM contains Bayesian regression, ridge regression, elastic net, and lasso estimator which is calculated by coordinate descent and least angle regression. We implemented stochastic gradient descent (SGD) algorithm for training GLM model. For our experiments, the proposed prediction model will be implemented using RStudio software with the caret library package and the H2O package for the prediction and regression algorithms.
2.5 Generation of the prediction model
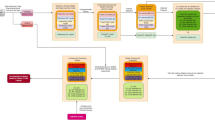
The main steps of the proposed deep neural network (DNN)–based prediction model are shown in Fig. 1. First, a DNN model generates twenty preliminary prediction models by applying training dataset and random hyperparameter values via the grid search algorithm [31, 32]. The used DNN algorithm is a deep feedforward network [33], consisting of 20 input neurons and three hidden layers, and generates a model with all combinations of hyperparameter values via not “hand tuning” but random grid search (RGS). All nodes of different layers used activation functions to determine the result of the neural network like Yes or No. Activation functions are typically non-linear functions such as Sigmoid, Softmax, tanh, ReLU, and Leaky ReLU. We applied the ReLU activation function because of its effectiveness. It simply replaces the negative values with zero and positive values remain unchanged. Moreover, Adam optimizer which is a stochastic gradient–based optimizer was used for weights optimization. During the model learning and generation processes, early stopping and 10-fold cross-validation method [34, 35] were used to select the optimal number of iterations automatically. Second, the performances of the preliminary prediction models were evaluated with the validation dataset. Third, the best hyperparameter values are selected and the best prediction model is generated from it. Moreover, the proposed model is evaluated by applying test dataset to generalize the performance. Here the role of the test dataset is to serve as a new dataset that the prediction model has never been applied before. Lastly, various performance matrixes such as accuracy, sensitivity, specificity, and the area under the ROC curve (AUC) are applied to evaluate the performance of proposed model.
2.6 Performance measures
We applied validation and test datasets to evaluate the accuracy of the MACE occurrence prediction models, and the experimental results will be described in different matrixes including accuracy, sensitivity, specificity, and the area under the ROC curve (AUC) for actual results versus predicted results.
2.7 Statistical analysis
Continuous variables (i.e., age and BP) were analyzed using a t test, and categorical variables (i.e., gender, DM, and smoking) were done using the chi-square test. Statistical significance is set to p value < 0.05. All the statistical analysis and data processing of all datasets were implemented using R software version 3.4.0.
3 Results
3.1 Baseline characteristics
After pre-processing the population, 10,813 AMI patients were selected for the experimental data, and the basic information of participating patients appeared in Table 2. The results showed that the occurrence of AMI was higher in young males than in females. Both also had higher pain, more STEMI in males, and higher dyspnea and Killip class in females than males. Clinical findings were higher in AMI patients than in healthy controls, especially in patients with AMI, CK, maximum CK-MB, maximum TnI, maximum TnT, NT-proBNP, and BNP. In the case of male’s medical history, smoking history, family history of heart disease, and complications were high, whereas previous angina before MI symptom, hypertension, and DM in female was higher than in male. In the medical procedure, thrombolysis and PCI performed much more in males than females.
3.2 Variable significance in prediction models
The significance of all variables in each prediction model was calculated as a percentage. The significance degree of the variables varies between 0 and 1: the significance of the most instrumental variable is 1 and the significance of the lowest is 0. Table 3 showed the top eight instrumental variables that each prediction model needs to predict the MACE occurrence during the 1, 6, and 12-month follow-up duration after discharge in AMI patients. The primary risk factors in the prediction model were different: (1) complications, CABG, pain, and history of DM in the deep learning model and (2) complications, taking a statin, NT-proBNP and angiographic findings in the GBM model, as well as (3) NT-proBNP, history of DM, and family history of HD in the GLM model. The machine learning–based models appeared a common variable for the attribute “complications” at 1 and 6-month follow-up after discharge, and the remaining models except for GLM did an instrumental variable for the attribute “age” at the 12-month. Note that the primary risk factors were different depending on the model and their significance was not always related to the accuracy of prediction models. Furthermore, the primary risk factors in machine learning–based models were also very different from those in traditional regression-based models.
3.3 Comparisons of AUC in prediction models
To evaluate the prediction models of the MACE occurrences in AMI patients, we compared the performance of prediction models according to the accuracy, sensitivity, specificity, and AUC. Table 4 showed the performance of all prediction models according to the evaluation indicators. The accuracy of all prediction models except the GRACE model was the highest at 1-month follow-up after discharge, and the DNN model showed more than 95% accuracy at 1, 6, and 12 months. Sensitivity showed the best value at the 12-month forecast in GBM model, and specificity was more than 95% in all models. Note that the DNN model showed the highest AUC value at 1-month follow-up after discharge, the GBM at 6-month, and the DNN and GBM models at 12-month.
4 Discussion
This paper proposed a deep learning–based prediction model of the MACE occurrences during the 1, 6, and 12-month follow-up after discharge in AMI patients and our results could be summarized as follows. This paper was the first trial of deep learning–based prediction model of the MACE occurrence. DNN method is a very successful technique that follows the same training method as the human brain for the prediction of the relationship between input data and target data. First, the primary risk factors in machine/deep learning models were CABG, complications (especially complications with DM), and pain. The attribute “age” in other models except GLM was the most instrumental variable in the prediction of MACE occurrences at 12-month follow-up. Second, the AUC values of MACE occurrence during the 1, 6, and 12-month follow-up after discharge were (1 M 0.97, 6 M 0.94, 12 M 0.96) in DNN and (0.96, 0.95, 0.96) in GBM. Consequently, the prediction accuracy in machine learning–based models was significantly superior to those of GRACE (0.75, 0.72, 0.76) in the regression-based model. The DNN model also showed more than 95% of accuracy at 1, 6, and 12-month follow-up after discharge.
It showed that machine/deep learning–based prediction models were suitable for the prediction of the MACE occurrences at 1, 6, and 12-month follow-up after discharge, and especially the prediction accuracies in machine/deep learning–based prediction models were superior to the established regression-based prediction model. The deep learning–based prediction model highly improved the prediction accuracy by automatically determining the primary risk factors required for MACE prediction in real time according to the characteristics of input data. In this paper, we found new primary risk factors for the prediction model of the MACE occurrences during 1, 6, and 12-month follow-up after discharge instead of traditional risk factors, such as smoking history, high blood pressure, high blood cholesterol, diabetes, physical inactivity, being overweight or obesity, family history of heart disease, and chest pain. As a result, the deep learning–based prediction model showed that it was not necessary to find the significant risk factors for the prediction model of MACE occurrences in AMI patients. The proposed model outperformed all the other machine learning and proved to be more successful for the prediction of the MACE occurrence in AMI clinical data.
There were some potential limitations in our research. First, KAMIR is not representative of all the world AMI patients because it is only a collection of Korean AMI patients. Second, the experimental data can be biased because the registered dataset does not have the information about the details of dosage, dose, and duration of taking drugs and beta blockers in each patient, as well as be limited to the short period follow-up of 1 year after hospital discharge.
5 Conclusion
This paper was the first deep learning–based prediction model of MACE occurrences during 1-year follow-up after discharge in AMI clinical data and highly improved the accuracy of prediction on KAMIR dataset using the knowledge and personal computing. Compared with a traditional GRACE risk prediction tool, the prediction accuracy of machine/deep learning–based prediction model was significantly higher. Especially, the proposed deep learning–based prediction model showed the accuracy of 95% or higher at 1, 6, and 12-month follow-up after discharge. Consequently, the deep learning approach was expected to provide a more efficient diagnosis and prediction tool for the MACE occurrences in AMI patients as a result of the knowledge-based personal computation in the future.
References
World Health Organization, The top 10 causes of death, 2017 [accessed Nov., 03, 2017], http://www.who.int/en/
Mahmood SS, Levy D, Vasan RS, Wang TJ (2014) The Framingham heart study and the epidemiology of cardiovascular disease: a historical perspective. Lancet 383(9921):999–1008. https://doi.org/10.1016/S0140-6736(13)61752-3
D’Agostino RB, Vasan RS, Pencina MJ, Wolf PA, Cobain M, Massaro JM, Kannel WB (2008) General cardiovascular risk profile for use in primary care: the Framingham heart study. Circulation 117(6):743–753. https://doi.org/10.1161/CIRCULATIONAHA.107.699579
Ferket BS, van Kempen BJH, Hunink MGM, Agarwal I, Kavousi M, Franco OH, Steyerberg EW, Max W, Fleischmann KE (2014) Predictive value of updating Framingham risk scores with novel risk markers in the US general population. PLoS One 9(2):e88312. https://doi.org/10.1371/journal.pone.0088312
Brindle P, Lampe F, Walker M, Whincup P, Fahey T, Ebrahim S (2003) Predictive accuracy of the Framingham coronary risk score in British men: prospective cohort study. BMJ 327(7426):1–6. https://doi.org/10.1136/bmj.327.7426.1267
Hippisley-Cox J, Coupland C, Vinogradova Y, Robso J, Minhas R, Sheikh A, Brindle P (2008) Predicting cardiovascular risk in England and Wales: prospective derivation and validation of QRISK2. BMJ 336(7659):1475–1482. https://doi.org/10.1136/bmj.39609.449676.25
Hippisley-Cox J, Coupland C, Brindle P (2017) Development and validation of QRISK3 risk prediction algorithms to estimate future risk of cardiovascular disease: prospective cohort study. BMJ 357:j2099. https://doi.org/10.1136/bmj.j2099
Antman EM, Cohen M, Bernink PJLM, McCabe CH, Horacek T, Papuchis G, Mautner B, Corbalan R, Radley D, Braunwald E (2000) The TIMI risk score for unstable angina/non-ST elevation MI: a method for prognostication and therapeutic decision making. JAMA 284(7):835–842. https://doi.org/10.1001/jama.284.7.835
Amin ST, Morrow DA, Braunwald E, Sloan S, Contant C, Murphy S, Antman EM (2013) Dynamic TIMI risk score for STEMI. J Am Heart Assoc 2(1):e003269. https://doi.org/10.1161/JAHA.112.003269
Fox KAA, Dabbous OH, Goldberg RJ, Pieper KS, Eagle KA, de Werf FV, Avezum Á, Goodman SG, Flather MD, Anderson FA Jr, Granger CB, for the GRACE Investigators (2006) Prediction of risk of death and myocardial infarction in the six months after presentation with acute coronary syndrome: prospective multinational observational study (GRACE). BMJ 333(7578):1–6. https://doi.org/10.1136/bmj.38985.646481.55
Elbarouni B, Goodman SG, Yan RT, Welsh RC, Kornder JM, Deyoung JP, Wong GC, Rose B, Grondin FR, Gallo R, Tan M, Casanova A, Eagle KA, Yan AT (2009) Validation of the Global Registry of Acute Coronary Event (GRACE) risk score for in-hospital mortality in patients with acute coronary syndrome in Canada. Am Heart J 158(3):392–399. https://doi.org/10.1016/j.ahj.2009.06.010
Huang W, FitzGerald G, Goldberg RJ, Gore J, McManus RH, Awad H, Waring ME, Allison J, Saczynski JS, Kiefe CI, Fox KAA, Anderson FA, McManus DD, TRACE-CORE Investigators (2016) Performance of the GRACE risk score 2.0 simplified algorithm for predicting 1-year death after hospitalization for an acute coronary syndrome in a contemporary multiracial cohort. Am J Cardiol 118(8):1105–1110. https://doi.org/10.1016/j.amjcard.2016.07.029
Mollee JS, Middelweerd A, Kurvers RL, Klein MCA (2017) What technological features are used in smartphone apps that promote physical activity? A review and content analysis. Pers Ubiquit Comput 21:633–643. https://doi.org/10.1007/s00779-017-1023-3
Spanakis G, Weiss G, Boh B, Lemmens L, Roefs A (2017) Machine learning techniques in eating behavior e-coaching. Pers Ubiquit Comput 21:645–659. https://doi.org/10.1007/s00779-017-1022-4
Law MR, Morris JK, Wald NJ (2009) Use of blood pressure lowering drugs in the prevention of cardiovascular disease: meta-analysis of 147 randomised trials in the context of expectations from prospective epidemiological studies. BMJ 338:b1665. https://doi.org/10.1136/bmj.b1665
Shetty A, Naik C (2016) Different data mining approaches for predicting heart disease. Int J Innov Res Sci Eng Technol 5(9):277–281. https://doi.org/10.15680/IJIRSET.2016.0505545
Mokashi AR, Tambe MN, Walke PT (2016) Heart disease prediction using ANN and improved K-means. Int J Innov Res Electr Electron Instrum Control Eng 4(4):221–224. https://doi.org/10.17148/IJIREEICE.2016.4454
Al-Maqaleh BM, Abdullah AMG (2016) An intelligent and electronic system based classification and prediction for heart disease diagnosis. Int J Emerg Trends Sci Technol 3(05):3951–3963. https://doi.org/10.18535/ijetst/v3i05.17
Subha V, Revathi M, Murugan D (2015) Comparative analysis of support vector machine ensembles for heart disease prediction. Int J Comput Sci Commun Netw 5(6):386–390
Ahmed A, Hannan SA (2012) Data mining techniques to find out heart diseases: an overview. Int J Innov Technol Explor Eng 1(4):18–23
Saqlain M, Wahid H, Saqib AS, Muazzam AK (2016) Identification of heart failure by using unstructured data of cardiac patient. In: 45th International conference on parallel processing workshops, pp 426–432. https://doi.org/10.1109/ICPPW.2016.66
Obermeyer Z, Emanue EJ (2016) Predicting the future—big data, machine learning, and clinical medicine. N Engl J Med 375(13):1216–1219
Ridker PM, Danielson E, Fonseca FAH, Genest J, Gotto AM, Kastelein JJP, Koenig W, Libby P, Lorenzatti AJ, MacFadyen JG, Nordestgaard BG, Shepherd J, Willerson JT, Glynn RJ (2008) Rosuvastatin to prevent vascular events in men and women with elevated C-reactive protein. N Engl J Med 359(21):2195–2207. https://doi.org/10.1056/NEJMoa0807646
Berglund E, Lytsy P, Westerling R (2013) Adherence to and beliefs in lipid-lowering medical treatments: a structural equation modeling approach including the necessity-concern framework. Patient Educ Couns 91(1):105–112. https://doi.org/10.1016/j.pec.2012.11.001
Wolterink JM, Leiner T, Viergever MA, Išgum I (2016) Dilated convolutional neural networks for cardiovascular MR segmentation in congenital heart disease, RAMBO 2016, HVSMR 2016: reconstruction, segmentation, and analysis of medical images 95–102. https://doi.org/10.1007/978-3-319-52280-7_9
Kook HY, Jeong MH, Oh S, Yoo SH, Kim EJ, Ahn YK, Kim JH, Chai LS, Kim YJ, Kim CJ, Cho MC (2014) Current trend of acute myocardial infarction in Korea (from the Korea Acute Myocardial Infarction Registry from 2006 to 2013). Am J Cardiol 114(12):1817–1822. https://doi.org/10.1016/j.amjcard.2014.09.019
Schmidhuber J (2015) Deep learning in neural networks: an overview. Neural Netw 61:85–117. https://doi.org/10.1016/j.neunet.2014.09.003
Friedman JH (2001) Greedy function approximation: a gradient boosting machine. Ann Stat 29(5):1189–1232 http://www.jstor.org/stable/2699986
Madsen H, Thyregod P (2010) Introduction to general and generalized linear models. CRC Press
Saqlain M, Jargalsaikhan B, Lee JY (2019) A voting ensemble classifier for wafer map defect patterns identification in semiconductor manufacturing. IEEE Trans Semicond Manuf 32(2):171–182. https://doi.org/10.1109/TSM.2019.2904306
Claesen M, Moor BD (2015) Hyperparameter search in machine learning, CoRR abs/1502.02127, pp 1–5
Bergstra J, Bengio Y (2012) Random search for hyper-parameter optimization. J Mach Learn Res:281–305
Goodfellow I, Bengio Y, Courville A (2016) Deep learning (adaptive computation and machine learning series), ch6. In: Deep feedforward networks, adaptive computation and machine learning series, pp 164–223
van den Broek EL, van der Sluis F, Dijkstra T (2013) Cross-validation of bimodal health-related stress assessment. Pers Ubiquit Comput 17:215–227. https://doi.org/10.1007/s00779-011-0468-z
Stone M (1974) Cross-validatory choice and assessment of statistical predictions. J R Stat Soc 36(2):111–147
Funding
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2017R1D1A1A02018718).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Kim, Y.J., Saqlian, M. & Lee, J.Y. Deep learning–based prediction model of occurrences of major adverse cardiac events during 1-year follow-up after hospital discharge in patients with AMI using knowledge mining. Pers Ubiquit Comput 26, 259–267 (2022). https://doi.org/10.1007/s00779-019-01248-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00779-019-01248-7