Abstract
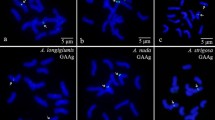
We applied fluorescent in situ hybridization (FISH) using (GAA)10 microsatellite repeat alone or in combination with oligo pSc119.2-1, pTa535-1, pAs1-1, 5S and 45S rDNA probes to identify individual chromosomes and produce banded karyotypes for five allopolyploid Aegilops species that were thought to possess M subgenome. Labelling patterns of GAA repeats allow identification of individual chromosomes in the studied species including diploid Ae. comosa subsp. heldreichii (2n = MhMh) and Ae. umbellulata (2n = UU); and allopolyploid Ae. biuncialis (2n = UbUbMbMb), Ae. geniculata (2n = UgUgMgMg), Ae. columnaris (2n = UcUcXcXc), Ae. neglecta (2n = UnUnXnXn) and Ae. crassa (2n = 4x = D1D1XcrXcr and 2n = 6x = D1D1XcrXcrDcrDcr). Intraspecific polymorphisms of GAA labelling patterns were observed between different genotypes in each species. FISH results showed that the X* subgenomes of Ae. columnaris and Ae. neglecta are highly similar, implying that both species originated from a common ancestor. However, these X* subgenomes were also distinct from and more heterochromatic when compared to the M* subgenomes of Ae. biuncialis and Ae. geniculata. This suggests that the X* subgenomes may not have originated from the Ae. comosa, as previously thought. Although (GAA)n bands are relatively few and weak in Ae. tauschii (DD genome), all 4x Ae. crassa (D1D1XcrXcr genome) chromosomes showed distinct GAA patterns. (GAA)10 probe alone was not sufficient for the discrimination of all the chromosomes of 6x Ae. crassa (D1D1XcrXcrDcrDcr), which needs a combination of (GAA)10 with the pTa535-1 or pAs1-1 probes.







Similar content being viewed by others
References
Appels R, Eversole K, Feuillet C, Keller B, Rogers J, Stein N, Pozniak CJ, Choulet F, Distelfeld A, Poland J (2018) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361:eaar7191. https://doi.org/10.1126/science.aar7191
Ardalani S, Mirzaghaderi G, Badakhshan H (2016) A Robertsonian translocation from Thinopyrum bessarabicum into bread wheat confers high iron and zinc contents. Pl Breed 135:286–290. https://doi.org/10.1111/pbr.12359
Badaeva ED, Friebe B, Gill BS (1996) Genome differentiation in Aegilops. 1. Distribution of highly repetitive DNA sequences on chromosomes of diploid species. Genome 39:293–306. https://doi.org/10.1139/g96-040
Badaeva E, Friebe B, Zoshchuk S, Zelenin A, Gill B (1998) Molecular cytogenetic analysis of tetraploid and hexaploid Aegilops crassa. Chromosome Res 6:629–637. https://doi.org/10.1023/A:1009257527391
Badaeva E, Zelenin A, Chikida N, Filatenko A (1999) Comparative analysis of the M genome in Aegilops comosa and Ae. heldreichii by means of C-banding technique and in situ hybridization. Russ J Genet 35:670–677
Badaeva E, Amosova A, Muravenko O, Samatadze T, Chikida N, Zelenin A, Friebe B, Gill B (2002) Genome differentiation in Aegilops. 3. Evolution of the D-genome cluster. Pl Syst Evol 231:163–190. https://doi.org/10.1007/s006060200018
Badaeva E, Amosova A, Samatadze T, Zoshchuk S, Shostak N, Chikida N, Zelenin A, Raupp W, Friebe B, Gill B (2004) Genome differentiation in Aegilops. 4. Evolution of the U-genome cluster. Pl Syst Evol 246:45–76. https://doi.org/10.1007/s00606-003-0072-4
Badaeva ED, Ruban A, Shishkina AA, Sibikeev SN, Druzhin AE, Surzhikov SA, Dragovich AY (2018) Genetic classification of Aegilops columnaris Zhuk. (2n = 4x = 28, UcUcXcXc) chromosomes based on FISH analysis and substitution patterns in common wheat × Ae. columnaris introgressive lines. Genome 61:131–143. https://doi.org/10.1139/gen-2017-0186
Bedbrook J, Jones J, O’dell M, Thompson R, Flavell R, Thompson R, Flavell R (1980) A molecular description of telomeric heterochromatin in Secale species. Cell 19:545–560. https://doi.org/10.1016/0092-8674(80)90529-2
Bernhardt N, Brassac J, Kilian B, Blattner FR (2017) Dated tribe-wide whole chloroplast genome phylogeny indicates recurrent hybridizations within Triticeae. BMC Evol Biol 17:141. https://doi.org/10.1186/s12862-017-0989-9
Chennaveeraiah MS (1960) Karyomorphologic and cytotaxonomic studies in Aegilops. Acta Horti Gothob 23:85–178
Cuadrado A, Schwarzacher T, Jouve N (2000) Identification of different chromatin classes in wheat using in situ hybridization with simple sequence repeat oligonucleotides. Theor Appl Genet 101:711–717. https://doi.org/10.1007/s001220051535
Cuadrado A, Cardoso M, Jouve N (2008) Increasing the physical markers of wheat chromosomes using SSRs as FISH probes. Genome 51:809–815. https://doi.org/10.1139/G08-065
Danilova TV, Akhunova AR, Akhunov ED, Friebe B, Gill BS (2017) Major structural genomic alterations can be associated with hybrid speciation in Aegilops markgrafii (Triticeae). Pl J 92:317–330. https://doi.org/10.1111/tpj.13657
Dennis E, Gerlach W, Peacock W (1980) Identical polypyrimidine-polypurine satellite DNAs in wheat and barley. Heredity 44:349–366
Dvorak J (1998) Genome analysis in the Triticum-Aegilops alliance. In: Slinkard AE (ed) Proceedings of 9th International Wheat Genetics Symposium, 2–7 August 1998, Saskatoon, Saskatchewan, Canada. University Extension Press, Saskatchewan, pp 8–11
Edet OU, Gorafi YS, Nasuda S, Tsujimoto H (2018) DArTseq-based analysis of genomic relationships among species of tribe Triticeae. Sci Rep 8:16397. https://doi.org/10.1038/s41598-018-34811-y
Eig A (1929) Monographisch-Kritische Uebersicht der Gatung Aegilops. In: Fedde F (ed) Repertorium specierum novarum regni vegetabilis. Verlag des Repertoriums, Dahlem bei Berlin, pp 1–228
Feldman M, Levy AA (2015) Origin and evolution of wheat and related triticeae species. In: Molnár-Láng M, Ceoloni C, Doležel J (eds) Alien introgression in wheat. Springer, Cham, pp 21–76
Flavell R, Bennett M, Smith J, Smith D (1974) Genome size and the proportion of repeated nucleotide sequence DNA in plants. Biochem Genet 12:257–269. https://doi.org/10.1007/BF00485947
Francki MG (2001) Identification of Bilby, a diverged centromeric Ty1-copia retrotransposon family from cereal rye (Secale cereale L.). Genome 44:266–274. https://doi.org/10.1139/g00-112
Friebe B, Jiang J, Tuleen N, Gill B (1995) Standard karyotype of Triticum umbellulatum and the characterization of derived chromosome addition and translocation lines in common wheat. Theor Appl Genet 90:150–156. https://doi.org/10.1007/BF00221010
Friebe B, Badaeva E, Kammer K, Gill B (1996) Standard karyotypes of Aegilops uniaristata, Ae. mutica, Ae. comosa subspecies comosa and heldreichii (Poaceae). Pl Syst Evol 202:199–210. https://doi.org/10.1007/BF00983382
Friebe BR, Tuleen NA, Gill BS (1999) Development and identification of a complete set of Triticum aestivum-Aegilops geniculata chromosome addition lines. Genome 42:374–380. https://doi.org/10.1139/g99-011
Friebe B, Zhang P, Linc G, Gill B (2005) Robertsonian translocations in wheat arise by centric misdivision of univalents at anaphase I and rejoining of broken centromeres during interkinesis of meiosis II. Cytogenet Genome Res 109:293–297. https://doi.org/10.1159/000082412
Fu S, Chen L, Wang Y, Li M, Yang Z, Qiu L, Yan B, Ren Z, Tang Z (2015) Oligonucleotide probes for ND-FISH analysis to identify rye and wheat chromosomes. Sci Rep 5:10552. https://doi.org/10.1038/srep10552
Gerlach W, Bedbrook J (1979) Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucl Acids Res 7:1869–1885
Gerlach W, Dyer T (1980) Sequence organization of the repeating units in the nucleus of wheat which contain 5S rRNA genes. Nucl Acids Res 8:4851–4865
Gerlach W, Peacock W (1980) Chromosomal locations of highly repeated DNA sequences in wheat. Heredity 44:269
Gill BS, Friebe B, Endo TR (1991) Standard karyotype and nomenclature system for description of chromosome bands and structural aberrations in wheat (Triticum aestivum). Genome 34:830–839. https://doi.org/10.1139/g91-128
Hammer K (1980) Vorarbeiten zur monographischen Darstellung von Wildpflanzensortimenten: Aegilops L. Kulturpflanze 28:33–180
Heslop-Harrison JS, Schwarzacher T (2011) Organisation of the plant genome in chromosomes. Pl J 66:18–33. https://doi.org/10.1111/j.1365-313X.2011.04544.x
Jaubert HF, Spach E (1853) Illustrationes Plantarum Orien-Talium, vol 4. Apud Roret Bibliopolam, Paris
Kato A, Lamb JC, Birchler JA (2004) Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc Natl Acad Sci USA 101:13554–13559. https://doi.org/10.1073/pnas.0403659101
Kihara H (1937) Genomanalyse bei Triticum und Aegilops VII. Kurze ubersicht uber die Ergebnisse der Jahre 1934–36. Mem Coll Agric Kyoto Univ 41:1–61
Kihara H (1949) Genomanalyse bei Triticum und Aegilops IX. Cytologia 14:135–144. https://doi.org/10.1508/cytologia.14.135
Kihara H (1954) Considerations on the evolution and distribution of Aegilops species based on the analyzer-method. Cytologia 19:336–357. https://doi.org/10.1508/cytologia.19.336
Kilian B, Mammen K, Millet E, Sharma R, Graner A, Salamini F, Hammer K, Ozkan H (2011) Aegilops. In: Kole C (ed) Wild crop relatives: Genomics and breeding resources, Cereals. Springer, Berlin, Heidelberg, pp 1–76
Kimber G, Feldman M (1987) Wild wheat, an Introduction. College of Agriculture University of Missouri, Columbia
Kimber G, Tsunewaki K (1988) Genome symbols and plasma types in the wheat group. In: Miller TE, Koebner RMD (eds) Proceedings of the 7th International Wheat Genetics Symposium, Cambridge, UK. Institute of Plant Science Research, Cambridge Laboratory, Cambridge, pp 1209–1210
Kimber G, Zhao YH (1983) The D genome of the Triticeae. Canad J Genet Cytol 25:581–589. https://doi.org/10.1139/g83-088
Komuro S, Endo R, Shikata K, Kato A (2013) Genomic and chromosomal distribution patterns of various repeated DNA sequences in wheat revealed by a fluorescence in situ hybridization procedure. Genome 56:131–137. https://doi.org/10.1139/gen-2013-0003
Levan A, Fredga K, Sandberg AA (1964) Nomenclature for centromeric position on chromosomes. Hereditas 52:201–220. https://doi.org/10.1111/j.1601-5223.1964.tb01953.x
Li S-F, Su T, Cheng G-Q, Wang B-X, Li X, Deng C-L, Gao W-J (2017) Chromosome evolution in connection with repetitive sequences and epigenetics in plants. Genes 8:290. https://doi.org/10.3390/genes8100290
Linc G, Gaál E, Molnár I, Icsó D, Badaeva E, Molnár-Láng M (2017) Molecular cytogenetic (FISH) and genome analysis of diploid wheatgrasses and their phylogenetic relationship. PLoS ONE 12:e0173623. https://doi.org/10.1371/journal.pone.0173623
Lukaszewski AJ (1997) Further manipulation by centric misdivision of the 1RS. 1BL translocation in wheat. Euphytica 94:257–261. https://doi.org/10.1023/A:1002916323085
Majka M, Kwiatek MT, Majka J, Wiśniewska H (2017) Aegilops tauschii accessions with geographically diverse origin show differences in chromosome organization and polymorphism of molecular markers linked to leaf rust and powdery mildew resistance genes. Frontiers Pl Sci 8:15. https://doi.org/10.3389/fpls.2017.01149
McIntyre C, Pereira S, Moran L, Appels R (1990) New Secale cereale (rye) DNA derivatives for the detection of rye chromosome segments in wheat. Genome 33:635–640. https://doi.org/10.1139/g90-094
Mehrotra S, Goyal V (2014) Repetitive sequences in plant nuclear DNA: types, distribution, evolution and function. Genom Proteom Bioinf 12:164–171. https://doi.org/10.1016/j.gpb.2014.07.003
Mirzaghaderi G, Marzangi K (2015) IdeoKar: an ideogram constructing and karyotype analyzing software. Caryologia 68:31–35. https://doi.org/10.1080/00087114.2014.998526
Mirzaghaderi G, Mason AS (2017) Revisiting pivotal-differential genome evolution in wheat. Trends Pl Sci 22:674–684. https://doi.org/10.1016/j.tplants.2017.06.003
Mirzaghaderi G, Mason AS (2019) Broadening the bread wheat D genome. Theor Appl Genet 132:1295–1307. https://doi.org/10.1007/s00122-019-03299-z
Mirzaghaderi G, Houben A, Badaeva ED (2014) Molecular-cytogenetic analysis of Aegilops triuncialis and identification of its chromosomes in the background of wheat. Molec Cytogenet 7:91. https://doi.org/10.1186/s13039-014-0091-6
Mirzaghaderi G, Abdolmalaki Z, Zohouri M, Moradi Z, Mason AS (2017) Dynamic nucleolar activity in wheat × Aegilops hybrids: evidence of C-genome dominance. Pl Cell Rep 36:1277–1285. https://doi.org/10.1007/s00299-017-2152-x
Molnar I, Vrana J, Buresova V, Capal P, Farkas A, Darko E, Cseh A, Kubalakova M, Molnar-Lang M, Dolezel J (2016) Dissecting the U, M, S and C genomes of wild relatives of bread wheat (Aegilops spp.) into chromosomes and exploring their synteny with wheat. Pl J 88:452–467. https://doi.org/10.1111/tpj.13266
Molnár I, Schneider A, Molnár-Láng M (2005) Demonstration of Aegilops biuncialis chromosomes in a wheat background by genomic in situ hybridization (GISH) and identification of U chromosomes by FISH using GAA sequences. Cereal Res Commun 33:673–680
Molnár I, Cifuentes M, Schneider A, Benavente E, Molnár-Láng M (2011a) Association between simple sequence repeat-rich chromosome regions and intergenomic translocation breakpoints in natural populations of allopolyploid wild wheats. Ann Bot (Oxford) 107:65–76. https://doi.org/10.1093/aob/mcq215
Molnár I, Kubaláková M, Šimková H, Cseh A, Molnár-Láng M, Doležel J (2011b) Chromosome isolation by flow sorting in Aegilops umbellulata and Ae. comosa and their allotetraploid hybrids Ae. biuncialis and Ae. geniculata. PLoS ONE 6:e27708. https://doi.org/10.1371/journal.pone.0027708
Molnár-Láng M, Ceoloni C, Doležel J (2015) Alien introgression in wheat: cytogenetics, molecular biology, and genomics. Springer International Publishing, Cham
Nagaki K, Tsujimoto H, Isono K, Sasakuma T (1995) Molecular characterization of a tandem repeat, Afa family, and its distribution among Triticeae. Genome 38:479–486. https://doi.org/10.1139/g95-063
Paszko B (2006) A critical review and a new proposal of karyotype asymmetry indices. Pl Syst Evol 258:39–48. https://doi.org/10.1007/s00606-005-0389-2
Pedersen C, Langridge P (1997) Identification of the entire chromosome complement of bread wheat by two-colour FISH. Genome 40:589–593. https://doi.org/10.1139/g97-077
Pedersen C, Rasmussen S, Linde-Laursen I (1996) Genome and chromosome identification in cultivated barley and related species of the Triticeae (Poaceae) by in situ hybridization with the GAA-satellite sequence. Genome 39:93–104. https://doi.org/10.1139/g96-013
Rayburn AL, Gill BS (1986) Isolation of a D-genome specific repeated DNA sequence from Aegilops squarrosa. Pl Molec Biol Reporter 4:102–109
Resta P, Zhang HB, Dubkovsky J, Dvořák J (1996) The origin of the genomes of Triticum biunciale, T. ovatum, T. neglectum, T. columnare, and T. rectum based on variation in repeated nucleotide sequences. Amer J Bot 83:1556–1565. https://doi.org/10.1002/j.1537-2197.1996.tb12813.x
Romero-Zarco C (1986) A new method for estimating karyotype asymmetry. Taxon 35:526–530. https://doi.org/10.2307/1221906
Ruban AS, Badaeva ED (2018) Evolution of the S-Genomes in Triticum-Aegilops alliance: evidences from chromosome analysis. Frontiers Pl Sci 9:1756. https://doi.org/10.3389/fpls.2018.01756
Ryan B, Joiner B (2001) Minitab handbook. Duxbury Resource Press, California
Schneider A, Molnár I, Molnár-Láng M (2008) Utilisation of Aegilops (goatgrass) species to widen the genetic diversity of cultivated wheat. Euphytica 163:1–19. https://doi.org/10.1007/s10681-007-9624-y
Schneider A, Molnár I, Molnár-Láng M, Benavente E, Cifuentes M (2010) Association between simple sequence repeat-rich chromosome regions and intergenomic translocation breakpoints in natural populations of allopolyploid wild wheats. Ann Bot (Oxford) 107:65–76. https://doi.org/10.1093/aob/mcq215
Serrato-Capuchina A, Matute D (2018) The role of transposable elements in speciation. Genes 9:254. https://doi.org/10.3390/genes9050254
Stebbins GL (1971) Chromosomal evolution in higher plants. Edward Arnold (Publishers) Ltd, London
Talbert LE, Kimber G, Magyar GM, Buchanan CB (1993) Repetitive DNA variation and pivotal-differential evolution of wild wheats. Genome 36:14–20. https://doi.org/10.1139/g93-003
Tang Z, Yang Z, Fu S (2014) Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc119.2, pTa-535, pTa71, CCS1, and pAWRC.1 for FISH analysis. J Appl Genet 55:313–318. https://doi.org/10.1007/s13353-014-0215-z
Tang S, Tang Z, Qiu L, Yang Z, Li G, Lang T, Zhu W, Zhhang J, Fu S (2018) Developing new Oligo probes to distinguish specific chromosomal segments and the A, B, D genomes of wheat (Triticum aestivum L.) using ND-FISH. Frontiers Pl Sci 9:1104
Teoh SB, Miller TE, Reader SM (1983) Intraspecific variation in C-banded chromosomes of Aegilops comosa and Ae. speltoides. Theor Appl Genet 65:343–348. https://doi.org/10.1007/BF00276575
Van Slageren MW (1994) Wild wheats: a monograph of Aegilops L. and Amblyopyrum (Jaub. & Spach) Eig (Poaceae). Agric Univ Wageningen Pap 94:37–380
Vishnyakova KS, Badaeva ED, Zelenin AV (1997) Study of intraspecific polymorphism of the C-banding patterns of Aegilops umbellulata L. chromosomes. Russ J Genet 33:623–627
Wang J, Zhang W, Zhao H, Li FR, Wang ZG, Ji J, Zhang XQ, Wang DW, Li JM (2013) Molecular cytogenetic characterization of the Aegilops biuncialis karyotype. Genet Molec Res 12:683–692. https://doi.org/10.4238/2013.March.11.16
Witcombe JR (1983) A guide to the species of Aegilops, L. Their taxonomy, morphology and distribution. IBPGR Secretariat, Rome
Zhang HB, Dvořák J (1992) The genome origin and evolution of hexaploid Triticum crassum and Triticum syriacum determined from variation in repeated nucleotide sequences. Genome 35:806–814. https://doi.org/10.1139/g92-123
Zhang P, Dundas IS, McIntosh RA, Xu SS, Park RF, Gill BS, Friebe B (2015) Wheat-Aegilops Introgressions. In: Molnár-Láng M, Ceoloni C, Doležel J (eds) Alien introgression in wheat. Springer, Switzerland, pp 221–243
Zhao YH, Kimber G (1984) New hybrids with D-genome wheat relatives. Genetics 106:509–515
Zhao L, Ning S, Yi Y, Zhang L, Yuan Z, Wang J, Zheng Y, Hao M, Liu D (2018) Fluorescence in situ hybridization karyotyping reveals the presence of two distinct genomes in the taxon Aegilops tauschii. BMC Genom 19:3. https://doi.org/10.1186/s12864-017-4384-0
Zhukovsky PM (1928) A critical-systematical survey of the species of the genus Aegilops L. Bull Appl Bot Genet Pl Breed 18:417–609
Zohary D, Feldman M (1962) Hybridization between amphidiploids and the evolution of polyploids in the wheat (Aegilops-Triticum) group. Evolution 16:44–61. https://doi.org/10.1111/j.1558-5646.1962.tb03197.x
Acknowledgements
Aegilops genotypes, as indicated in Table 1, were kindly provided by Dr. W. J. Raupp and Dr. B. S. Gill, Wheat Genetic & Genomic Resources Center, Kansas State University, USA or provided by the IPK gene bank, Germany. This work was originally the MSc thesis of ZA at the University of Kurdistan. GM was supported by a fellowship within a 3-month DAAD program to visit ASM at Justus Liebig University in 2018. ASM is supported by DFG Emmy Noether grant MA6473/1-1.
Author information
Authors and Affiliations
Contributions
GM conceived and designed research. ZA and GM conducted experiments. ASM contributed to manuscript revisions and critical discussions. EB contributed to chromosome identification, critical discussions and manuscript revisions.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Handling editor: Martin A. Lysak.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Abdolmalaki, Z., Mirzaghaderi, G., Mason, A.S. et al. Molecular cytogenetic analysis reveals evolutionary relationships between polyploid Aegilops species. Plant Syst Evol 305, 459–475 (2019). https://doi.org/10.1007/s00606-019-01585-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-019-01585-3