Abstract
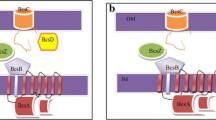
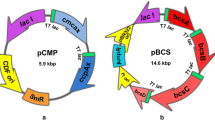
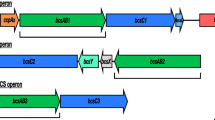
Based on cellulose biosynthesis pathway of Gluconacetobacterxylinus BPR2001 and E. coli Nissle 1917, bcsA and bcsB genes have been selected and bioinformatics studies done to the analyses of nucleotide and amino acid sequence alignment, stability of RNA, protein, and promotor power. We amplify and clone bcsA, bcsB, and bcsAB genes of G. xylinus BPR2001 in Escherichiacoli Nissle 1917 under the inducible tac promoter. Our results of bioinformatics predictions demonstrate similar active site and three-dimensional structure of BcsA and BcsB proteins in two different bacteria. In addition, our data reveal that BcsA and BcsB proteins of E. coli have weaker promotor power, RNA secondary structure, and protein stability than that of the same proteins in G. xylinus. Some of the reasons of BcsAB protein selection from G. xylinus and its heterologous expression in E. coli is the noted points. Production of the related proteins visualized using SDS-PAGE. We find out that Congo red absorbance at 490 nm has no significant difference in wild-type strain (E. coli Nissle 1917) compared to recombinants bcsA+ or bcsB+, but recombinant bcsAB+ could produce more cellulose than that of the wild-type strain. Furthermore, the measurement of cellulose dry weights of all samples confirms bacterial cellulose production enhancement in recombinant bcsAB+ (1.94 g l−1). The FTIR analysis reveals that the crystallinity indices do not change significantly after over expressing each of genes.







Similar content being viewed by others
References
Ebrahimi E, Babaeipour V, Khanchezar S (2016) Effect of down-stream processing parameters on the mechanical properties of bacterial cellulose. Iran Polym J. https://doi.org/10.1007/s13726-016-0462
Meftahi A, Nasrollahi D, Babaeipour V, Alibakhshi S, Sahbazi S (2015) Investigation of nano bacterial cellulose coated by sesamum oil for wound dressing application. Proced Mater Sci. https://doi.org/10.1016/j.mspro.2015.11.109
Römling U (2002) Molecular biology of cellulose production in bacteria. Res Microbiol. https://doi.org/10.1016/S0923-2508(02)01316-5
Lu H, Jiang X (2014) Structure and properties of bacterial cellulose produced using a trickling bed reactor. Appl Biochem Biotechnol. https://doi.org/10.1007/s12010-014-0795-4
Kawano S, Tajima K, Uemori Y, Yamashita H, Erata T, Munekata M, Takai M (2002) Cloning of cellulose synthesis related genes from Acetobacter xylinum ATCC23769 and ATCC53582: comparison of cellulose synthetic ability between strains. DNA Res. https://doi.org/10.1093/dnares/9.5.149
Lee KY, Buldum G, Mantalari A, Bismarck A (2014) More than meets the eye in bacterial cellulose: biosynthesis, bioprocessing, and applications in advanced fiber composites. Macromol Biosci. https://doi.org/10.1002/mabi.201300298
Omadjela O, Narahari A, Strumillo J, Mélid H, Mazur O, Bulone V, Zimmer J (2013) BcsA and BcsB form the catalytically active core of bacterial cellulose synthase sufficient for in vitro cellulose synthesssis. Proc Natl Acad Sci USA 110:17856–17861
Saxena IM, Kudlicka K, Okuda K, Brown JRM (1994) Characterization of genes in the cellulose-synthesizing operon (acs operon) of Acetobacter xylinum: implications for cellulose crystallization. J Bacteriol 176:5735–5752
Tal R, Wong HC, Calhoon R, Gelfand D, Fear AL, Volman G, Mayer R, Ross P, Amikam D, Weinhouse H, Cohen A, Sapir S, Ohana P, Benziman M (1998) Three cdg operons control cellular turnover of cyclic di-GMP in Acetobacter xylinum: genetic organization and occurrence of conserved domains in isoenzymes. J Bacteriol 180:4416–4425
Zogaj X, Nimtz M, Rohde M, Bokranz W, Römling U (2001) The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol Microbiol. https://doi.org/10.1046/j.1365-2958.2001.02337.x
Takai MM (2002) Cloning of cellulose synthesis related genes from Acetobacter xylinum ATCC23769 and ATCC53582: comparison of cellulose synthetic ability between strains. DNA Res. https://doi.org/10.1093/dnares/9.5.149
Römling U, Galperin MY (2015) Bacterial cellulose biosynthesis: diversity of operons, subunits, products, and functions. Trends Microbiol. https://doi.org/10.1016/j.tim.2015.05.005
Sajadi E, Babaipour V, Deldar AA, Yakhchali B, Fatemi SSA (2017) Enhancement of crystallinity of cellulose produced by Escherichia coli through heterologous expression of bcsD gene from Gluconacetobacter xylinus. Biotechnol Lett. https://doi.org/10.1007/s10529-017-2366-6
Römling U, Gomelsky M, Galperin MY (2005) C-di-GMP: the dawning of a novel bacterial signaling system. Mol Microbiol. https://doi.org/10.1111/j.1365-2958.2005.04697.x
Amikam D, Galperin MY (2006) PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics. https://doi.org/10.1093/bioinformatics/bti739
Fritsch E, Sambrook J (2012) Molecular cloning: a laboratory manual, 4th edn. Cold Spring Harbor Laboratory Press, New York, pp 94–109
Seo EJ, Weibel S, Wehkamp J, Oelschlaeger TA (2012) Construction of recombinant E. coli Nissle 1917 (EcN) strains for the expression and secretion of defensins. Int J Med Microbiol. https://doi.org/10.1016/j.ijmm.2012.05.002
Pulicherla KK, Kumar A, Gadupudi GS, Kotra SR, Rao KR (2013) In vitro characterization of a multifunctional staphylokinase variant with reduced reocclusion, produced from salt inducible E. coli GJ1158. Biomed Res Int. https://doi.org/10.1155/2013/297305
Da Re S, Ghigo JM (2006) A CsgD-independent pathway for cellulose production and biofilm formation in Escherichia coli. J Bacteriol. https://doi.org/10.1128/JB.188.8.3073-3087.2006
Toyosaki H, Naritomi T, Seto A, Matsuoka M, Tsuchida T, Yoshinaga F (1995) Screening of bacterial cellulose producing Acetobacter strains suitable for agitation culture. Biosci Biotechnol Biochem. https://doi.org/10.1271/bbb.59.1498
Nobles DR, Brown R (2008) Transgenic expression of Gluconacetobacter xylinus strain ATCC 53582 cellulose synthase genes in the cyanobacterium Synechococcus leopoliensis strain UTCC 100. Cellulose. https://doi.org/10.1007/s10570-008-9217-5
Nobles DR, Brown R (2007) Many paths up the mountain: tracking the evolution of cellulose biosynthesis in cellulose. Mol Struct Biol. https://doi.org/10.1007/978-1-4020-5380-1_1
Ryjenkov DA, Simm R, Romling U, Gomelsky M (2006) The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J Biol Chem. https://doi.org/10.1074/jbc.C600179200
Lin FC, Brown RMJ (1989) Purification of cellulose synthase from Acetobacterxylinum. In: Schuerch C (ed) Cellulose and wood chemistry and technology. Wiley, New York, pp 473–492
Lee VT, Matewish JM, Kessler JL, Hyodo M, Hayakawa Y, Lory S (2007) A cyclic-di-GMP receptor required for bacterial exo polysaccharide production. Mol Microbiol. https://doi.org/10.1111/j.1365-2958.2007.05879.x
Wong HC, Feara L, Calhoon RD, Eichinger GH, Mayer R, Amikam D, Benziman M, Gelfand DH, Meade JH, Emerick AW (1990) Genetic organization of the cellulose synthase operon in Acetobacter xylinum. Proc Natl Acad Sci 87:8130–8134. https://doi.org/10.1073/pnas.87.20.8130
Kumar A, Negi YS, Choudhary V, Bhardwaj NK (2014) Characterization of cellulose nanocrystals produced by acid-hydrolysis from sugarcane bagasse as agro-waste. J Nonlinear Optic Phys. https://doi.org/10.12691/jmpc-2-1-1
Sessler TH (2014) Bacterial cellulose in cyanobacteria: enhancement of cellulose production in Synechococcus elongatus with Gluconacetobacter xylinus transgenes. Dissertation, University of Texas at Austin
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All of authors declare that they do not have any conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sajadi, E., Fatemi, S.SA., Babaeipour, V. et al. Increased cellulose production by heterologous expression of bcsA and B genes from Gluconacetobacterxylinus in E. coli Nissle 1917. Bioprocess Biosyst Eng 42, 2023–2034 (2019). https://doi.org/10.1007/s00449-019-02197-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-019-02197-4