Abstract
Peanut, found to be relatively drought tolerant crop, has been the choice of study to characterize the genes expressed under gradual water deficit stress. Nearly 700 genes were identified to be enriched in subtractive cDNA library from gradual process of drought stress adaptation. Further, expression of the drought inducible genes related to various signaling components and gene sets involved in protecting cellular function has been described based on dot blot experiments. Fifty genes (25 regulators and 25 functional related genes) selected based on dot blot experiments were tested for their stress responsiveness using northern blot analysis and confirmed their nature of differential regulation under different field capacity of drought stress treatments. ESTs generated from this subtracted cDNA library offered a rich source of stress-related genes including signaling components. Additional 50% uncharacterized sequences are noteworthy. Insights gained from this study would provide the foundation for further studies to understand the question of how peanut plants are able to adapt to naturally occurring harsh drought conditions. At present functional validation cannot be deemed in peanut, hence as a proof of concept seven orthologues of drought induced genes of peanut have been silenced in heterologous N. benthamiana system, using virus induced gene silencing method. These results point out the functional importance for HSP70 gene and key regulators such as Jumonji in drought stress response.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Peanut or groundnut (Arachis hypogaea L.) is the fourth most important oil seed in the world, cultivated mainly in tropical, subtropical and warm temperate climates (FAO 2004). It adapts to wide range of environments. It is cultivated in about 8 million hectares in India. Once established, rainfall of 500 to 1,000 mm will allow commercial production of peanut, although crop can be produced on as little as 300–400 mm of rainfall. Recent physiological studies provide hints that peanut is a relatively drought tolerant crop having improved water use efficiency mechanisms (Nautiyal et al. 2002). Drought is one of the most important abiotic stresses that cause adverse effects on the productivity of crops (Boyer 1982). Its multigenic, incompletely penetrant, quantitative nature makes it difficult to breed for drought tolerance. In response to environmental extremes plants have developed several adaptive mechanisms, which allow them to survive adverse conditions. The evolved adaptive mechanisms are displayed by different degrees of tolerance, largely determined by their genetic plasticity.
One of the major molecular responses that plants exhibit to drought stress is altered expression of genes, related to different pathways associated with stress perception, signal transduction, regulators and synthesis of a number of compounds (Ramanjulu and Bartels 2002; Sreenivasulu et al. 2007). Several hundred genes that respond to drought stress at the transcriptional level have been identified in model crop Arabidopsis by microarray technology and other means (Seki et al. 2002; Shinozaki and Yamaguchi-Shinozaki 2007). The adaptive mechanisms under stress are a net effect of altered cell metabolism resulting from regulated expression of stress responsive genes. The resurrection plants have better capabilities to cope with severe drought conditions; hence, several studies have been conducted to discover what key genes are involved in enabling these plants to survive desiccation. The molecular aspects of desiccation tolerance in resurrection plants such as Craterostigma plantagineum (Bartels et al. 1990; Bartels and Salamini 2001; Phillips et al. 2002; Bartels 2005), Xerophyta viscosa (Mundree et al. 2000; Mowla et al. 2002; Dahlia et al. 2003), Xerophyta humilis (Collett et al. 2003), Sporobolus stapfianus (Neale et al. 2000) and Selaginella lepidophylla (Iturriaga et al. 2000) reveal complex mechanisms of desiccation tolerance (Bernacchia and Furini 2004).
Although some mechanisms of stress response are common to all cells, there exist major differences in strategies adopted by plants to cope with desiccation stress (Ramanjulu and Bartels 2002; Smith-Espinoza et al. 2003). Realizing this, it would be more rewarding to explore crop species with higher levels of stress tolerance at molecular level. Evidences support the fact that stress responsive genes from tolerant species provide better protection to cellular structures due to existence of genes that code for structurally and/or functionally efficient stress proteins associated with stress adaptation (Waditee et al. 2002; Majee et al. 2004; Dastidar et al. 2006). There is tremendous amount of biological diversity among different plant species that necessitates sampling of other plant genome sequences, to understand the diversity of gene content and basic functional resolution of the plant genomes in general. To identify stress specific genes, it would be more rewarding to isolate the differentially expressed genes providing a clear picture of the transcriptome under stress from relatively drought tolerant crop. There are a number of approaches to identify the differentially expressed genes and to enrich stress-responsive genes from model crop species whose genome size is relative large. These include differential display (Liang and Pardee 1992; Cho et al. 2001), subtractive hybridization, suppressive subtractive hybridization (Diatchenko et al. 1996), cDNA-AFLP (Kivioja et al. 2005) etc.
The main objective of this study was to identify, isolate and characterize the genes expressed during gradual drought stress acclimation in peanut. At present it is difficult to find answers for important questions like how plants such as peanut are able to cope with drought stress. The first step in this quest, as well as in many other biological investigations, is to construct a cDNA library enriched for differentially expressed transcripts, and eventually to annotate them to gain preliminary insights of drought tolerance. In this direction, a cDNA library enriched for peanut genes expressed specifically in response to gradual drought stress was constructed and determined for their DNA sequences to functionally annotate these drought stress induced expressed sequence tags (ESTs). Characterization of their stress responsive nature indicated that many of the genes isolated are involved in drought stress response. For subset of drought induced peanut genes, its orthologues were identified in N. benthamiana and functionally validated them by VIGS approach. These results further confirm their potential involvement in water deficit stress tolerance.
Materials and methods
Plant growth conditions and drought stress imposition
The seeds of all crop species was procured form the University of Agricultural Science (UAS) farm, Bangalore. The plants were grown, with recommended dose of fertilizer and appropriate prophylactic measures. Plants were cultivated at 12 h light/12 h dark cycles with maximum temperature of 28°C by mid day and minimum temperature of 18°C during night with light intensity reaching maximum of 1,000–1,200 μmol m−2 s−1. The amount of water the soil can hold is expressed as mass percentage, and it is considered as 100% field capacity (FC) of soil. Twenty-five days old peanut plants, maintained at 100% FC were stressed by decreasing the soil water status gradually (decreasing water status by approximately 10% FC/day). Plants were maintained at specific FC (100, 80, 70, 60, 50, 40, 30 and 20% FC) for 5 days, and stress was monitored by gravimetrically weighing the pots twice a day. Fully expanded leaves were collected, at the end of stress period, from both stressed and non-stressed plants (100% FC) for RNA isolation and leaf water relation studies. At the time of collecting leaf samples, the exact soil water potential was calculated using a WP4 dewpoint potentiometer (Decagon Devices Inc., Washington, DC, USA). At 40% FC, the soil water potential was −0.73 MPa and at 20% FC it was −1.22 MPa, confirming that the plants were stressed. Relative water content (RWC), leaf solute potential, osmotic adjustment and electrolyte leakage were analysed as a measure of leaf water relation.
Leaf water relations
The RWC was calculated as percentage, as described by Barrs and Weatherly (1986); leaf solute potential of leaf sap was estimated as described by Govind (2006) using vapour pressure osmometer (VAPRO) (Wescor Inc., Logan, UT, USA). The values obtained in mmols Kg−1 was converted to MPa and calculated ψS = −mols Kg−1 (RK), where R = 0.008314 and K = 2950. ψS100 = (ψs × RWC)/100. Using RWC and solute potential, extent of osmotic adjustment in leaves was derived (Flower and Ludlow 1986; Subbarao et al. 2000) which were in osmotic adjustment (OA) = drought leaf ψS100 − irrigated leaf ψS100. Electrolyte leakage was estimated following protocol modified from Tripathy et al. (2000), using conductivity meter (Elico-India, CM183, EC-TDS analyser). The loss of membrane integrity was determined as percentage leakage.
Assessing stress damage using seedling survival test, leaf area retention, chlorophyll damage in leaf disc assay
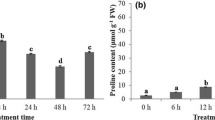
For seedling survival test, 1-week-old seedlings were stressed by withholding water for 3 days and allowed to alleviate at the end of stress for 48 h. The percentage of seedlings surviving was recorded. Leaf area retention test was carried out for 20-day-old plants. Stress was imposed by withholding irrigation for 1 week and alleviated at the end of stress for 48 h. Leaf area was measured using leaf area meter (Delta-T, Delta T Devices Ltd, England) and expressed as percent leaf area retention compared to the control plants (Fig. 1a). Ten leaf discs from fully expanded leaves of 25-day-old plants were soaked in either water or PEG (−1.8 MPa) or NaCl (400 mM) for 48 h under continuous light of 350 μEm-2s-1 for leaf disc assay. Pigments were isolated using acetone:DMSO (1:1) mix and absorbance recorded at 553 nm to estimate pheophytin (Govind 2006). For all the experiments, five replications were maintained, and the data was subjected to two-tailed t test at 5% level of significance.
Screening different crop species for a seedling survival and leaf area retention after drought stress; b chlorophyll damage in leaf discs exposed to drought and salinity. Mean values and standard error was calculated from five independent experiments. Within each set of experiment, bars with different letters were significantly different at 0.05% level
Peanut subtracted drought stress cDNA library construction—Modified SMART kit protocol
RNA isolated from freshly collected leaves maintained at 100% FC was used as driver and 40–20% FC (−0.73 to −1.22 MPa) pooled sample as tester in subtraction. Total RNA was isolated from plant materials using Trizol reagent (Invitrogen, Carlsbad, CA, USA), according to the manufacture’s instructions. The quantity and quality of total RNA was evaluated by spectrophotometry (OD260/280) and formaldehyde–1% agarose gel electrophoresis. cDNA library was constructed using the SMART cDNA synthesis kit in pTriplEx2 (Clontech, Palo Alto, CA, USA). To perform subtraction using this kit, primers for driver were synthesized in such a way that the sequence of the primers was randomized without altering the GC content and the forward primer was biotinylated, and for tester the primers provided in the kit was used. First strand of driver and tester was carried out using kit protocol. The driver cDNA pool was amplified using oligo dT and biotinylated Gcap amplification primers. The biotinylated strand was captured using streptavidin coated magnetic beads after denaturing double stranded DNA at 95°C for 5 min and by immediately cooling on ice. Single stranded tester cDNA was allowed to hybridize with biotinylated driver cDNA pool at 65°C for 3 h. Two rounds of hybridizations were carried out to normalize and subtract driver transcripts. After placing the tubes in a magnetic separator, the stress specific pool was isolated. The resulting cDNA was packed into λ phages using the Gigapack III Gold packaging kit (Stratagene, La Jolla, CA, USA). The pTriplEx2 phagemid clones in Escherichia coli were obtained using the mass in vivo excision protocol according to the manufacture’s instructions (Clontech, USA). The white clones grown on screening LB medium (Carbencillin/IPTG/X-Gal) were recovered by random colony selection.
-
Primers used:
-
Driver: Reverse primer: 5′agatgctgacgagtcgagtagcgccgc(t)303′
Forward primer: 5′gaacgatgctagcgtaagtccagtcagtgaacggccggg3′ biotnylated at 5′
-
Tester: Reverse primer: 5′aagcagtggtctagaaccaagtggccattacggccggg3′
Forward primer: 5′attcgagctctagaggccgaggcggccgacatg(t)303′
Dot blot/reverse northern analysis
Only clones with insert size of 400 bp or higher were used for expression study. PCR product (2 ng) of recombinant clones and vector control (negative control) was blotted onto nylon membrane and cross-linked using UV. Two such membranes were probed with labelled control and stress cDNA, respectively. Labelling was carried out during first strand cDNA synthesis, primed by oligo dT, using MMLV reverse transcriptase. Hybridization was carried out at 60°C, and signals captured on a phosphoimager. RNA isolated from plants maintained at 100% FC and 40–20% FC (pooled sample) were used to prepare control and stress probes, respectively. The blots were quantitatively scored using phosphoimager. The resulted expression values were normalized using median centring normalization method according to Sreenivasulu et al. (2006) and heat maps were developed for the same.
Sequencing and annotation
Only clones found to be stress responsive by dot blot analysis were sequenced. Approximately 200 ng/μl of the column purified plasmid was used for sequencing using ABI prism. The vector sequences were trimmed and the resulted clean EST sequences were subjected to annotation using NCBI database—BlastX (translated query vs. protein database) against non-redundant SWISS PROT database.
Northern/RNA blot analysis
Northern blot hybridizations were carried out according to Sambrook and Russell (2001). RNA (15 μg) from each sample was separated in formaldehyde denaturing gel and transferred to Hybond nylon membrane and fixed by UV (1,200 μJ for 60 s) in a UV cross-linker. The blots were probed with respective inserts, prepared by labelling with [32P] dCTP (3,000 Ci mmol−1) during PCR. Pre-hybridization was carried out at 42°C for 2 h and hybridization at 60°C overnight with blocking solution (0.5 M sodium–phosphate buffer, pH 7.2, 1 mM EDTA and 7% SDS). High stringent washes were carried out according to Sambrook and Russell (2001). Ouvrard group kindly provided the known stress responsive genes, which were shown in their study to be stress responsive (Ouvrard et al. 1996). The blots were exposed to phosphoimager plate for 2 days; the intensity of band was quantified and normalized for variation in RNA loaded. The extent of expression under stress was presented as percent increase over control, considering expression under control as 100%. The expression was calculated using formula: (absolute value under stress × 100/absolute value under control) − 100.
Characterization of N. benthamiana VIGS plants
A few selected peanut water deficit induced genes were selected and identified their orthologous sequences from N. benthamiana and silenced them using VIGS approach (refer Senthil-Kumar et al. 2007). The control plants were maintained at 100% FC. Two sets of plants were maintained as controls, while one set of plants were mock treated [wild type, TRV2 alone (mock), the other set of plants with TRV2-gene was maintained under non-stress condition. For water deficit stress, three replications were maintained. Gradual stress was imposed by decreasing the soil water status over a period of 1 week to specific FC of 50% and maintained for 2 days at 50% FC. Water status was monitored gravimetrically by weighing the pots at regular intervals. At the end of stress, leaf samples were collected from both stressed and non-stressed plants. The extent of pheophytin accumulation under stress, as a measure of chlorophyll damage, was recorded by measuring absorbance of the pigments isolated using acetone: DMSO mix at 553 nm. Total chlorophyll was estimated, by recording absorbance at 663, 652 and 645 nm. Pheophytin to chlorophyll ratio was calculated as a measure of stress index. The results presented are from three independent experiments.
Results
Identifying peanut as relatively drought tolerant crop based on screening of several crop species
A number of crop species such as peanut, sunflower, cowpea, beans, horse gram, tomato, finger millet and maize were screened, for their extent of stress tolerance, at seedling and later stages of growth. Among the crop species tested for seedling survival, under drought stress imposition for 3 days, peanut, horse gram and finger millet were found to be most tolerant with nearly 90% survivability. On the contrary sunflower, beans and tomato were found to be highly susceptible to drought with only 20% survival rate (Fig. 1a). Similar trend was found among the crops species with respect to leaf area retention under stress. Further, we measured pheophytin content to estimate the chlorophyll degradation in leaf discs of 20-day-old plants from control and drought stress treatments (−1.8 MPa). While the pheophytin content has slightly increased in peanut, horse gram and fingermillet from 0.5 to 1.0 mg/g between control and drought stress conditions, we noticed a substantial increase of pheophytin content (up to 2.0 mg/g) in drought susceptible varieties such as sunflower, beans and tomato under drought stress (Fig. 1b). Based on these parameters, we designated peanut as relatively drought tolerant crop and considered for further molecular studies of drought tolerance.
Optimization of gradual water deficit stress induction protocol
Twenty days old peanut plants were subjected to water deficit stress. Stress maintained by gravimetric approach was confirmed by measuring soil water potential, where in soil ψ reached −1.22 MPa in soil maintained at 20% FC. Measurement of leaf osmotic potential and RWC, of plants subjected to different levels of soil FC, indicated that peanut plants reached critical RWC of 50% only at 20% FC. Leaf osmotic potential increased from 358 to 935 mmol/kg under 20% FC. Such an increase in osmotic potential may be explained to some extent by osmotic adjustment, increasing from 0.1007 at 60% FC to 0.377 at 20% FC. No drastic loss in membrane integrity was observed in plants exposed to severe stress, probably because of an alteration in cell metabolism under stress leading to maintenance of membrane integrity (Table 1). A few known stress responsive genes coding for ELIP, LEA2 and nsLTP known to be highly expressed under stress were used as probes, to identify the water deficit stress level required for maximum expression of stress genes in peanut. Northern analysis indicated that during the process of gradual water stress in peanut, maximum expression of stress genes occurs across a wide range of stress level between 40 and 20% FC (Fig. 2). Hence, genes differentially expressed under stress (40–20% FC) were isolated based on subtractive cDNA library and identified the genes induced by gradual water deficit stress, which probably play role in imparting tolerance under water deficit.
Isolation and characterization of gradual water deficit stress induced genes in peanut
During library construction the cDNA subtraction hybridization procedure was employed between mRNA populations of drought treated (pooled 40–20% FC) and control (100% FC) samples (see “Materials and methods” for further details) to normalize and subtract control transcripts. As a result the mRNA pool enriched for water deficit treatments from peanut plants were isolated and the resulted cDNA library (in house naming: AhWSI—Arachis hypogaea water stress induced library) not only represent drought induced gene set but also ensured elimination of constitutively expressed abundant transcripts. By doing so, we have successfully eliminated housekeeping and non-target genes. The clones were further screened by dot blot to confirm stress responsive nature of these clones. More than 700 clones are found to be drought-responsive in peanut. Normalization of expression data obtained from two replicated experiments was used to verify reproducibility. Average expression ratio was calculated between control and drought stress treatments, and subjected the log transformed expression values for producing heat maps as described earlier according to Sreenivasulu et al. (2006).
All the stress responsive clones validated from dot blot experiments were sequenced to generate peanut water stress induced ESTs. Upon sequencing and quality check performance, nearly 500 sequences have been deposited in NCBI gene bank (Acc. No. EC268400–EC268685; EC365167–EC365455). The library comprises many different categories of genes, encompassing many classes of gene products that are induced by water deficit. These genes can be classified into different classes based on their predicted protein function (Supplemental Table 1). Of the total 693 clones, functional categorization was carried out for 350 clones based on BLAST output with cut off score value of 50 (Supplemental Table 1, Fig. 3). The remaining clones were considered as unassigned or unknown with no similarity or with no BLAST hit result.
The overrepresented stress induced clones were broadly classified into several functional categories involved in (a) Metabolism: photosynthesis (4.3%), amino acid metabolism (2.3%), carbohydrate metabolism (3.4%), nucleic acid metabolism (1.4%), fatty acid metabolism (1.4%), metal handling related (2.9%), energy related (1.4%) and secondary metabolism (1.4%), (b) cellular processes: cell cycle (1.7%), protein synthesis (4.9%), post translational process related (4.9%), protein degradation (6.9%), (c) protecting cellular structures: biotic stress (3.1%), abiotic stress (11.7%), oxidative stress (2.0%) and (d) regulators: regulation of transcription (8.3%), hormone regulated (3.4%) (Supplemental Table 1, Fig. 3).
Among the genes isolated, major share was occupied by genes related to protecting cellular structures under abiotic stress (11.7%) followed by genes regulating transcription contributing nearly 8.3% and hormone/signaling related 6.8%. The gene sets mentioned in afore mentioned categories are discussed in detail. Heat maps were developed after quantifying and normalizing the intensities of expression from dot blot experiment for regulators as well for abiotic stress related transcripts depicted in Figs. 4 and 5. To further validate and study their regulation under different water stress, expression pattern of selected transcripts were studied in peanut plants maintained at different levels of soil water status by northern analysis. Obtained data also shed light into further details of how these categories of genes are regulated at different levels of drought stress (Figs. 6, 7).
Expression profiles of selected regulatory genes induced under drought stress from dot blot experiments were shown as heat maps. Expression values are given in logarithmically scaled (base 2) signal intensities: red high expression, yellow moderate expression, blue low expression. Clone identity, putative BLAST description and defined functional classes are provided on the right side of figure
Expression profiles of selected abiotic and biotic stress responsive genes induced under drought stress from dot blot experiments were shown as heat maps. Expression values are given in logarithmically scaled (base 2) signal intensities: red high expression, yellow moderate expression, blue low expression. Clone identity, putative BLAST description and defined functional classes are provided on the right side of figure
Levels of transcripts of AhWSI clones coding for putative regulatory sequences differentially accumulated under different levels of water deficit stress measured using Northern blot analysis method. The values obtained from control samples considered as 100% and respective induction or repression of expression under different levels of water deficit stress is calculated (see “Materials and methods” for further details) and indexed above northern blots
Levels of transcripts of AhWSI clones coding for putative functional proteins differentially accumulated under different levels of water deficit stress measured using northern blot analysis method. The values obtained from control samples considered as 100% and the respective induction or repression of expression under different levels of water deficit stress is calculated (see “Materials and methods” for further details) and indexed above northern blots
Drought induced regulatory genes
One of the main objective of the present study is identifying regulatory genes (includes transcription factors, kinases, phosphatases, hormone influenced and secondary messengers all contributing in signal perception and signal transduction) which are induced under water deficit treatments from drought tolerant legume plant, peanut. Many families of transcription factors including AP2/EREBP (AhWSI 279), bHLH (AhWSI 111, AhWSI 40), bZIP (AhWSI 20), CCAAT box (AhWSI 117), Homeobox (AhWSI6 11), Jumonji (AhWSI 72, AhWSI 116), NAC (AhWSI 153, AhWSI 308) and several zinc finger protein transcripts are preferentially induced under drought treatments in peanut plants (Fig. 4; Supplemental Table 2). Also among the upstream signaling components we observed induction of transcripts of calmodulins (AhWSI 227, AhWSI 228), G protein (AhWSI 551), MAPKK (AhWSI 28) and several receptor kinases during drought treatments. In addition, we also noticed specific upregulation of hormone responsive genes such as auxin-repressed proteins (AhWSI 306, AhWSI 468, AhWSI 467), brassinosteroid responsive BRH1 (AhWSI 36), cytokinin-repressed protein CR9 (AhWSI 465), GA like proteins (AhWSI 291, AhWSI 464) during drought treatments (Fig. 4; Supplemental Table 2).
Twenty-five clones coding for putative regulatory genes involved in signal perception, signal transduction and regulation of gene expression by transcription factors were considered to test their stress responsiveness under different FC of drought stress treatments. Of the 25 clones studied, 21 clones accumulated maximum transcripts under severe stress, and the remaining 5 clones (AhWSI 153, AhWSI 63, AhWSI 115, AhWSI 147, AhWSI 58) were highly expressed under moderate stress (60–40% FC) (Fig. 6). These include NAC transcription factor, enhancer protein, probable helicase, 5-hydroxy tryptamine receptor and CBS domain containing proteins which show high expression under moderate stress, but decline with increase in severity of stress. Transcription factors like DNA binding enhancer protein, SNF family protein and HTH transcription factor had maximum transcripts at severe stress of 20% FC. Most of the signaling associated genes showed an increase in expression with increase in severity of drought stress, except for hydroxyl tryptamine receptor. Though many regulatory genes are induced under stress to moderate level, here, we found some exceptional cases. Specifically among them, four genes Calcineurin like phosphoesterase family protein (AhWSI 17), TRF1 interacting kinase (AhWSI 60), DNA binding enhancer protein (AhWSI 63) and Zn finger protein (AhWSI 285), were found to be highly induced by stress, with transcripts level increasing more than 100–200% in comparison to their respective controls (Fig. 6).
Drought induced genes involved in protecting cellular function
The proteins encoded by this category of genes are basically involved in protecting cellular macromolecules and structures under stress. The most abundant in our library were those belonging to late embryogenesis abundant proteins, heat shock proteins, DnaJ like proteins, aldehyde reductase, proline rich protein and defensins, which are known for their stress responsive nature (Fig. 5; Supplemental Table 2). LEA proteins have considerable hydrophylicity and are known to have important function in protecting cells from damage under desiccation. Heat shock proteins and DnaJ like proteins function mainly as chaperones. Very little is known regarding the role of proline rich proteins under stress. Though defensins are majorly known to be upregulated in response to biotic stress, here, we noticed their induction under abiotic stress such as drought. Its functional relevance under drought is yet to be determined.
Additional twenty-five clones belongs to selected groups like abiotic stress related, metabolism associated, transporters and protein modification/chaperonins have been tested for their temporal regulation under gradual water deficit ranging from 60 to 20% FC. All the clones tested are induced under mild stress, and their expressions are either maintained under moderate or severe stress (Fig. 7). Among the broad category of abiotic stress regulated proteins, HSPs, Drought induced protein and LEAs were highly expressed when plants were subjected to moderate levels of stress (60% FC). Among the metabolism related ones, those associated with amino acid and lipid metabolism were highly upregulated under moderate stress. On the other hand carbon metabolism and secondary metabolism related genes were maintained high even under severe drought stress. Notably, genes coding for aldehyde reductase, salt induced protein and ferritin were upregulated under severe drought stress of 40–20% FC. Interestingly, the two transporters selected for the study displayed high levels of expression only under sever stress of 20% FC.
Characterization of selected N. benthamiana virus induced gene silencing (VIGS) plants
We have considered seven VIGS plants out of previously characterized 25 VIGS plants from N. benthamiana (Senthil-Kumar et al. 2007) based on the criteria that water deficit induced genes identified in the present study from peanut should have more than 80% nucleotide similarity with N. benthamiana. Functional relevance of these seven orthologous genes has been tested in N. benthamiana by measuring pheophytin content from control and drought stress treatments (Fig. 8) and discussed their relevance in the context of drought tolerance. This biochemical trait turned out to be useful in assessing stress tolerance index where we compared several species in our studies and provided evidence that peanut is a relatively drought tolerant crop (Fig. 1). Hence, we used this biochemical trait to characterize seven VIGS plants. These include silencing of drought induced Lea5, HSP20 and HSP70, which belongs to broad group of chaperone proteins, having protective function under stress. However, only HSP70 showed higher pheophytin levels under drought stress in comparison to mock treated plants (Fig. 8). Our results confirmed that HSP70 seems to play an important role in conferring drought tolerance. Besides, another most interesting aspect noticed is down-regulation of Jumonji (JMJC), bHLH, and Zinc finger regulatory genes which made the transgenic tobacco plants more tolerant to drought, and these silenced plants showed lower chlorophyll degradation under severe water deficit conditions. These results suggest that these are negative regulators, in particular JMJC having positive effect in conferring drought tolerance.
Discussion
Until the genome sequence information is available for all the crops, researchers have to rely on information generated by studying model crops and explore EST sequences from crop species (Ewing et al. 1999). There is overwhelming evidence that stress genes from adapted species are effective in the perception of stress signal and eventually trigger down stream structurally and functionally efficient proteins, shown to be involved in protecting cellular structures and molecules under stress (Waditee et al. 2002; Majee et al. 2004; Dastidar et al. 2006). Our physiological studies revealed that peanut is a relatively drought tolerant crop in comparison to sunflower, beans and tomato. In spite of drastic decrease in leaf water potential and RWC reaching critical levels of 50% under severe drought stress, the extent of chlorophyll degradation was less (Fig. 1). This could be because peanut plants are able to maintain turgor driven process as a result of osmotic adjustment, in turn leading to maintenance of cellular metabolism. There are a couple of drought stress cDNA library already generated in A. hypogaea, but they correspond to genes expressed in immature pods (Luo et al. 2005; Devaiah et al. 2007). There is one library, which catalogues the genes expressed in a wild Arachis species (A. stenosperma) under non-stress condition (Proite et al. 2007). In our study, a peanut subtracted drought stress cDNA library was constructed from late stages of plant development, i.e. 25-day-old plants exposed to gradual drought stress, mimicking stress occurring under field condition. Expression analysis of known stress responsive genes (ELIP, LEA2 and nsLTP) indicated that optimum stress could be imposed by the above mentioned stress protocol (Fig. 2), and this leads to maximum expression of stress genes during the process of gradual imposition of severe drought stress.
Though from model species like Arabidopsis, several regulatory genes have been found to be expressed within a few hours of exposure to stress during early stages of plant development (Abe et al. 2003; Kang et al. 2002), there is a dearth in knowledge of regulatory cascades operating during later stages of plant development exposed to abiotic stresses. One of the important tasks we chose to investigate in the present study is to gain preliminary insights about up-stream signaling related genes preferentially induced in drought tolerant peanut plant under water deficit. Expression analysis of regulatory genes revealed that most of these genes were stress responsive, showing diverse expression between moderate to severe stress. Among the drought induced gene set, we identified several proteins involved in the synthesis or perception of different hormones. Among them several auxin-repressed proteins (ARP) are preferentially induced under drought treatments (Fig. 4), indicating that drought stress response trigger ARP in peanut. Though many auxin-induced genes are reported from plants, ARP gene function in stress adaptation is less known. Interestingly, ARP genes are known to be preferentially expressed during pollen maturation (Steiner et al. 2003) and during strawberry fruit maturation (Reddy and Poovaiah 1990). Afore mentioned developmental processes are connected with natural desiccation mechanism. In this context, we propose that ARP might play an important role in desiccation tolerance in peanut. Also cytokinin-repressed CR9 and counteracting brassinosteroid-responsive BRH1 genes were induced under drought treatments.
Genes encoding several proteins involved in signaling cascades were also found to be induced under drought in peanut. These include calcium binding proteins (calmodulins), calcineurin and calcium dependent protein kinases (CDPK) (Figs. 4, 6), which are potentially induced due to altered free calcium levels in the cytosol as one of the earliest abiotic stress responses. Also serine/threonine protein kinase (Seki et al. 2001; Zhu 2002), phospholipase D (Maarouf et al. 1999; Wang and Wang 2001; Guo et al. 2006), cAMP binding protein (Pascual-Ahuir et al. 2001) are known to be induced under stress. In accordance, here, we observed the upregulation of several down stream signaling related MAPKK, receptor kinases as well as secondary messengers such as phospholipase D and rhodopsins which are preferentially induced under drought treatments (Figs. 4, 6).
In the present study, we also identified key transcription factor related genes differentially expressed under different water deficit conditions from relatively drought tolerant crop peanut (Figs. 4, 6; Supplemental Table 2). Among them of special interest is induction of two Jumonji transcription factors in drought treated plants. Recently, jumonji domain containing proteins have been shown to act as demethylases, shown to involve in chromatin structure and gene expression (Chen et al. 2006). The most highly represented TF families which are induced to drought in peanut are the zinc fingers, WRKY, AP2, Myb and NAC. Several members of these families were previously identified as being responsive to various stresses, which includes EREBP, Helicase (Gong et al. 2002, 2005; Sanan-Mishra et al. 2005; Owttrim 2006), bHLH (Abe et al. 2003; Wang et al. 2003; Li et al. 2006) and Zn finger protein (Dhundy et al. 1998; Cheong et al. 2002; Kreps et al. 2002; Mukhopadhyay et al. 2004; Davletova et al. 2005) (Fig. 6). In addition, many of the stress responsive genes identified from our present study have previously not been reported under stress, in plants. A few of them are reported to be involved in pathways usually occurring under non-optimal conditions of animal cells. For example, TRF2 (TTAGGG repeat binding factor 2), a Myb related protein is involved in protecting the telomere ends in humans (Broccoli et al. 1997; Agata et al. 2000). NCK2 interacting kinases are usually known for initiating downstream signaling pathways regulating cytoskeleton assembly and remodeling (Cowan and Henkemeyer 2002).
It is presumed that the genes expressed during the course of gradual stress in tolerant species are responsible for altering the cellular metabolism, leading to adaptation under severe stress. Among the well known examples are genes encoding proteins related to protection of cellular structures and denaturation of proteins and enzymes. These classes of proteins encode LEA/dehydrins; many of them are known to accumulate during dehydration response and also known to accumulate during embryo desiccation tolerance (Ingram and Bartels 1996; Sreenivasulu et al. 2006). Genes encoding these proteins are over-represented in our differential expressed gene set. Also in our drought-responsive ESTs, collection of large number of putative HSP encoding genes, which are known to act as molecular chaperones, proteins protecting against stress damage (Wang et al. 2004) are represented. In addition, other functional genes include ferritin (Lobréaux et al. 1993; Allen 1995; Goto et al. 2001), extensin (Yoshiba et al. 2001), peroxidase (Nakano and Asada 1981; Agnès et al. 2006), which are also found to be stress-responsive (Fig. 5). Taken together, peanut plants are able to regulate expression of genes known to impart tolerance by protecting macromolecules like membranes, proteins etc. and thereby maintaining optimal membrane integrity under drought. There are a number of genes in our library which are also expressed in wild relative of Arachis (A. stenosperma) for example PR proteins, ADH, proline rich proteins, metallothionien etc. Also among the overrepresented functional category of enriched sequences we noticed the expression of proteases and its inhibitors from drought treated peanut library (Supplemental Table 2).
Besides these well known genes, we observed preferential expression of aquaporin related PIP genes (AhWSI 304, AhWSI 305) under water deficit conditions (Supplemental Table 2), which play an important role in plant water relationship. Surprisingly, we also noticed induction of few transcripts related to photosynthesis such as RuBisCO-interacting proteins, chloroplast chaperones and components related to photosystem I and II under drought in peanut (Supplemental Table 2). Very few studies have documented the modulation of PSI under stress conditions. The PSI components are largely integrated and composed of many subunits making it energetically expensive for the cell to produce under stress. Hence, the over-expression of genes related to PSI components under drought could be part of integrated response of a tolerant plant to reduce photoinhibition as well as to maintain energy balance. Similar situation is reported with respect to enhanced expression of photosynthesis related transcripts for another drought tolerant horsegram plant (Reddy et al. 2008).
Taken together the results of our findings, we conclude that cDNA subtraction method applied here to enrich drought-responsive genes from drought tolerant peanut plant resulted in identifying several hundred genes coding for stress adaptation and complex signaling components. Notably, many of the drought-responsive genes (50%) coding for unknown genes identified from peanut plants did not showed significant similarity to Arabidopsis genome sequences. Thereby provide further platform to characterize functional relevance of these genes in the context of drought stress adaptation in near future. As of now we cannot deem of implementing VIGS system in peanut for functional validation due to the lack of efficient transformation system. Hence in the present study, we have chosen seven peanut drought induced genes, identified the corresponding orthologues which have been silenced in N. benthamiana using VIGS approach (see Senthil-Kumar et al. 2007) and estimated pheophytin content to measure tolerance index. Using this approach, we investigated the role of N. benthaminana Lea5, HSP20 and HSP70 genes involved in protecting cellular function and confirmed that only HSP70 seems to play potential role in drought tolerance. Although HSP20 and Lea5 have been shown to be responsive in the present study as well in other species (Galau et al. 1993), its suppression did not result in susceptibility. Another most interesting aspect is down-regulation of Jumonji (JMJC) made the transgenic tobacco plants more tolerant to drought. These silenced plants showed lower chlorophyll degradation under severe water deficit conditions suggesting that JMJC might act as negative regulators in imparting drought tolerance. These data, lead us to conclude that recruitment of HSP70 and JMJC in tolerance against drought might be conserved between dicotyledonous plant species such as peanut and tobacco and further demonstrates that VIGS is a useful approach to dissect tolerance for highly conserved genes in a genetically intractable plant species. In conclusion, our study suggests that peanut can effectively re-programme globally its complex signalling networks to activate regulated expression of several genes to mitigate the stress induced cellular damage. These results could also facilitate the understanding of cellular mechanisms involving groups of gene products that act in coordination in response to stimuli of water withhold.
Change history
19 November 2018
The original version of this article contains the following duplicate figures, namely blots showing expression data of clones.
Abbreviations
- AhWSI:
-
Arachishypogaea water stress induced library
- ARP:
-
Auxin repressed protein
- bHLH:
-
Basic helix loop helix protein
- BRH1:
-
Brassinosteroid-responsive
- EST:
-
Expressed sequence tag
- FC:
-
Field capacity
- HSP:
-
Heat shock protein
- JMJC:
-
Jumonji class
- Lea:
-
Late embryogenesis abundant protein
- FC:
-
Field capacity
- PDS:
-
Phytoene desaturase
- RWC:
-
Relative water content
- TRV:
-
Tobacco rattle virus
- VIGS:
-
Virus-induced gene silencing
References
Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15:63–78
Agata S, van Steensel B, Alessandro B, Stefan O, Matthias RS, Gisela S, de Lange T (2000) Control of human telomere length by TRF1 and TRF2. Mol Cell Biol 20(5):1659–1668
Agnès DL, Roselyne FI, Dominique CA, Pham-Thi AT, Zuily-Fodil Y (2006) Isolation and characterization of four ascorbate peroxidase cDNAs responsive to water deficit in Cowpea leaves. Ann Bot 97(1):133–140
Allen RD (1995) Dissection of oxidative stress tolerance using transgenic plants. Plant Physiol 107:1049–1054
Barrs HD, Weatherly PE (1986) A re-examination of the relative turgidity for estimating water deficits in leaves. Aust J Biol Sci 15:413–428
Bartels D (2005) Desiccation tolerance studied in the resurrection plant Craterostigma plantagineum. Integr Comp Biol 45:696–701
Bartels D, Salamini F (2001) Desiccation tolerance in the resurrection plant Craterostigma plantagineum: a contribution to the study of drought tolerance at the molecular level. Plant Physiol 127:1346–1353
Bartels D, Schneider K, Terstappen G, Piatkowski D, Salamini F (1990) Molecular cloning of ABA-modulated genes from the resurrection plant Craterostigma plantagineum which are induced during desiccation. Planta 181:27–34
Bernacchia G, Furini A (2004) Biochemical and molecular responses to water stress in resurrection plants. Physiol Plant 121:175–181
Boyer JS (1982) Plant productivity and environment. Science 281(4571):443–448
Broccoli D, Smogorzewska A, Chong L, de Lange T (1997) Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat Genet 17:231–235
Chen Z, Zang J, Whetstine J, Hong X, Davrazou F, Kutateladze TG, Simpson M, Mao Q, Pan CH, Dai S, Hagman J, Hansen K, Shi Y, Zhang G (2006) Structural insights into histone demethylation by JMJD2 family members. Cell 125:691–702
Cheong YH, Chang HS, Gupta R, Wang X, Zhu T, Luan S (2002) Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiol 129:661–677
Cho YJ, Meade JD, Walden JC, Chen X, Guo Z, Liang P (2001) Multicolor fluorescent differential display. Biotechniques 30:562–572
Collett H, Butowt R, Smith J, Farrant J, Illing N (2003) Photosynthetic genes are differentially transcribed during the dehydration–rehydration cycle in the resurrection plant, Xerophyta humilis. J Exp Bot 54:2593–2595
Cowan CA, Henkemeyer M (2002) Ephrins in reverse, park and drive. Trends Cell Biol 12:339–346
Dahlia G, Thomson JA, Mundree SG (2003) Molecular characterization of XVSAP1, a stress-responsive gene from the resurrection plant Xerophyta viscose Baker. J Exp Bot 54(381):191–201
Dastidar KG, Maitra S, Goswami L, Roy D, Das KP, Majumder AL (2006) An Insight into the molecular basis of salt tolerance of l-myo-inositol 1-P synthase (PcINO1) from Poteresia coarctata (Roxb.) Tateoka, a halophytic wild rice. Plant Physiol 140:1279–1296
Davletova S, Schlauch K, Coutu J, Mittler R (2005) The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiol 139:847–856
Devaiah KM, Bali G, Athmaram TN, Basha MS (2007) Identification of two new genes from drought tolerant peanut up-regulated in response to drought. Plant Growth Regul 52:249–258
Dhundy RB, Vaijaynti VP, Winicov I (1998) Alfin1, a novel zinc-finger protein in alfalfa roots that binds to promoter elements in the salt-inducible MsPRP2 gene. Plant Mol Biol 38(6):1123–1135
Diatchenko L, Lau YF, Campbell AP, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov ED, Siebert PD (1996) Suppression subtractive hybridization: a method for generating differentially regulated or tissue specific cDNA probes and libraries. Proc Natl Acad Sci USA 93:6025–6030
Ewing RM, Kahla AB, Poirot O, Lopez F, Audic S, Claverie JM (1999) Large-scale statistical analyses of rice ESTs reveal correlated patterns of gene expression. Genome Res 9:950–959
Flower DJ, Ludlow MM (1986) Contribution of osmotic adjustment to the dehydration tolerance of water-stressed pigeon pea [Cajanus cajan (L.) Millsp.] leaves. Plant Cell Environ 9:33–40
Galau G, Wang HYC, Hughes DW (1993) Cotton Lea5 and Lea14 encode atypical late embryogenesis-abundant proteins. Plant Physiol 101:695–696
Gong Z, Lee H, Xiong L, Jagendorf A, Stevenson B, Zhu JK (2002) RNA helicase-like protein as an early regulator of transcription factors for plant chilling and freezing tolerance. Proc Natl Acad Sci USA 99(17):11507–11512
Gong Z, Dong CH, Lee H, Zhu J, Xiong L, Gong D, Stevenson B, Zhu JK (2005) A DEAD box RNA Helicase is essential for mRNA export and important for development and stress responses in Arabidopsis. Plant Cell 17(1):256–267
Goto F, Yoshihara T, Masuda T, Takaiwa F (2001) Genetic improvement of iron content and stress adaptation in plants using ferritin gene. Biotechnol Genet Eng Rev 18:351–371
Govind G (2006) Profiling transcriptome of Arachis hypogaea L. under drought and monitoring the expression pattern, Ph.D. thesis, University of Agricultural Sciences (B), India
Guo BZ, Xu G, Cao YG, Holbrook CC, Lynch RE (2006) Identification and characterization of phospholipase D and its association with drought susceptibilities in peanut (Arachis hypogaea). Planta 223(3):512–520
Ingram J, Bartels D (1996) The molecular basis of dehydration tolerance in plants. Annu Rev Plant Physiol Plant Mol Biol 47:377–403
Iturriaga G, Gaff DF, Zentella R (2000) New desiccation-tolerant plants, including a grass, in the central highlands of Mexica, accumulate trehalose. Aus J Bot 48(2):153–158
Kang J, Choi H, Im M, Kim SY (2002) Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14:343–357
Kivioja T, Arvas M, Saloheimo M, Penttilä M, Ukkonen E (2005) Optimization of cDNA-AFLP experiments using genomic sequence data. Bioinformatics 21(11):2573–2579
Kreps JA, Wu Y, Chang HS, Zhu T, Wang X, Harper JF (2002) Transcriptome changes for Arabidopsis in response to salt, osmotic, and clod stress. Plant Physiol 130:2129–2141
Li X, Duan X, Jiang H, Sun Y, Tang Y, Yuan Z, Guo J, Liang W, Chen L, Yin J, Ma H, Wang J, Zhang D (2006) Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol 141(4):1167–1184
Liang P, Pardee AB (1992) Differential display of eukaryotic mesenger RNA by means of polymerase chain reaction. Science 257:967–971
Lobréaux S, Hardy T, Briat JF (1993) Abscisic acid is involved in the iron-induced synthesis of maize ferritin. EMBO J 12(2):651–657
Luo M, Dang P, Guo BZ, He G, Holbrook CC, Bausher MG, Lee RD (2005) Generation of expressed sequence tags (ESTs) for gene discovery and marker development in cultivated peanut. Crop Sci 45:346–353
Maarouf HE, Zuily-Fodil Y, Gareil M, d’Arcy-Lameta A, Pham Thi At (1999) Enzymatic activity and gene expression under water stress of phospholipase D in two cultivars of Vigna unguiculata L. Walp. differing in drought tolerance. Plant Mol Biol 39(6):1257–1265
Majee M, Maitra S, Dastidar KG, Pattnaik S, Chatterjee A, Hait NC, Das KP, Majumder AL (2004) A novel salt-tolerant l-myo-inositol-1-P synthase from Poteresia coarctata (Roxb.) Tateoka, a halophytic wild rice, molecular cloning, bacterial overexpression, characterization, and functional introgression into tobacco-conferring salt tolerance phenotyope. J Biol Chem 279: 28539–28552
Mowla SB, Thomson J, Farrant JM, Mundree SG (2002) A novel stress-inducible antioxidant enzyme identified from the resurrection plant Xerophyta viscosa baker. Planta 215:716–726
Mukhopadhyay A, Vij S, Tyagi AK (2004) Overexpression of a zinc-finger protein gene from rice confers tolerance to cold, dehydration, and salt stress in transgenic tobacco. Proc Natl Acad Sci USA 101(16):6309–6314
Mundree SG, Whittaker A, Thomson JA, Farrant JM (2000) An aldose reductase homolog from the resurrection plant Xerophyta viscosa Baker. Planta 211:693–700
Nakano Y, Asada K (1981) Hydrogen peroxide is scavenged by ascorbate-specific peroxidase in spinach chloroplasts. Plant Cell Physiol 22:867–880
Nautiyal PC, Rachaputi NR, Joshi YC (2002) Moisture-deficit-induced changes in leaf-water content, leaf carbon exchange rate and biomass production in groundnut cultivars differing in specific leaf area. Field Crop Res 74:67–79
Neale AD, Blomstedt CK, Bronson P, Le TN, Guthridge K, Evans J, Gaff DF, Hamill JD (2000) The isolation of genes from the resurrection grass Sporobolus stapfianus which are induced during severe drought stress. Plant Cell Environ 23:265–277
Ouvrard O, Cellier F, Ferrare K, Tousch D, Lamaze T, Dupuis JM, Casse-Delbart F (1996) Identification and expression of water stress- and abscisic acid-regulated genes in a drought tolerant sunflower genotype. Plant Mol Biol 31:819–829
Owttrim GW (2006) RNA helicases and abiotic stress. Nucleic Acids Res 34(11):3220–3230
Pascual-Ahuir A, Posas F, Serrano R, Proft M (2001) Multiple levels of control regulate the yeast cAMP-response element-binding protein repressor Sko1p in response to stress. J Biol Chem 276(40):37373–37378
Phillips JR, Oliver MJ, Bartels D (2002) Molecular genetics of desiccation and tolerant systems. In: Black M, Pritchard HW (eds) Desiccation and survival in plants: drying without dying. CAB International, Wallingford, pp 319–341
Proite K, Soraya CM Leal-Bertioli, David JB, Márcio CM, Felipe R da Silva, Natalia FM, Patrícia MG (2007) ESTs from a wild Arachis species for gene discovery and marker development. BMC Plant Biol 7: 7
Ramanjulu S, Bartels D (2002) Drought- and desiccation-induced modulation of gene expression in plants. Plant Cell Environ 25:141–151
Reddy ASN, Poovaiah BW (1990) Molecular cloning and sequencing of a cDNA for an auxin-repressed mRNA: correlation between fruit growth and repression of the auxin-regulated gene. Plant Mol Biol 14:127–136
Reddy PCO, Sairanganayakulu G, Thippeswamy M, Sudhakar Reddy P, Reddy MK, Sudhakar Chinta (2008) Identification of stress-induced genes from the drought tolerant semi-arid legume crop horsegram [Macrotyloma uniflorum (Lam.) Verdc.] through analysis of subtracted expressed sequence tags. Plant Sci 175:372–384
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Sanan-Mishra N, Pham XH, Sopory SK, Tuteja N (2005) Pea DNA helicase 45 overexpression in tobacco confers high salinity tolerance without affecting yield. Proc Natl Acad Sci USA 102(2):509–514
Seki M, Narusaka M, Abe H, Kasuga M, Yamaguchi-Shinozaki K, Carninic P, Hayashizaki Y, Shinozaki K (2001) Monitoring the expression pattern of 1,300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell 13:61–72
Seki M, Narusaka M, Kamiya A (2002) Functional annotation of a full-length Arabidopsis cDNA collection. Science 296:141–145
Senthil-Kumar M, Govind G, Kang L, Mysore KS, Udayakumar M (2007) Functional characterization of Nicotiana benthamiana homologs of peanut water deficit-induced genes by virus-induced gene silencing. Planta 225(3):523–539
Shinozaki K, Yamaguchi-Shinozaki K (2007) Gene networks involved in drought stress response and tolerance. J Exp Bot 58(2):221–227
Smith-Espinoza CJ, Richter A, Salamini F, Bartels D (2003) Dissecting the response to dehydration and salt (NaCl) in the resurrection plant Craterostigma Plantagenium. Plant Cell Environ 26:1307–1315
Sreenivasulu N, Radchuk V, Strickert M, Miersch O, Weschke W, Wobus U (2006) Gene expression patterns reveal tissue-specific signaling networks controlling programmed cell death and ABA-regulated maturation in developing barley seed. Plant J 47(2):310–327
Sreenivasulu N, Sopory SK, Kishor PBK (2007) Deciphering the regulatory mechanisms of abiotic stress tolerance in plants by genomic approaches. Gene 388(1–2):1–13
Steiner C, Bauer J, Amrhein N, Bucher M (2003) Two novel genes are differentially expressed during early germination of the male gametophyte of Nicotiana tabacum. Biochim Biophys Acta 1625:123–133
Subbarao GV, Chauhan YS, Johansen C (2000) Patterns of osmotic adjustment in pigeon pea: its importance as a mechanism of drought resistance. Eur J Agron 12:239–249
Tripathy JN, Zhang J, Robin S, Nguyen TT, Nguyen HT (2000) QTLs for cell-membrane stability mapped in rice (Oryza sativa L.) under drought stress. Theor Appl Genet 100:1197–1202
Waditee R, Hibino T, Nakamura T, Incharoensakdi A, Takabe T (2002) Over expression of Na+/H+ antiporter confers salt tolerance on fresh water Cyanobacterium, making it capable of growth in sea water. Proc Natl Acad Sci USA 99(6):4109–4114
Wang C, Wang X (2001) A novel phospholipase D of Arabidopsis that is activated by oleic acid and associated with the plasma membrane. Plant Physiol 127:1102–1112
Wang YJ, Zhang ZG, He XJ, Zhou HL, Wen YX, Dai JX, Zhang JS, Chen SY (2003) A rice transcription factor OsbHLH1 is involved in cold stress response. Theor Appl Genet 107(8):1402–1409
Wang WX, Vinocur B, Shoseyov O, Altman A (2004) Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci 9(5):244–252
FAO Statistical Yearbook (2004). http://www.fao.org/statistics/yearbook/vol_1_1/site_en.asp?page=production
Yoshiba Y, Aoki C, Iuchi S, Nanjo T, Seki M, Sekiguchi F, Yamaguchi-Shinozaki K, Shinozaki K (2001) Characterization of four extensin genes in Arabidopsis thaliana by differential gene expression under stress and non-stress conditions. DNA Res 8(3):115–122
Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53:247–273
Acknowledgments
Geetha Govind, Patricia J and Senthil Kumar M, thank the Council of Scientific and Industrial Research (CSIR), New Delhi, India for the financial support of Senior Research Fellowship award [No. 9/271(70)/2002/EMR-1, No. 9/271(84)/2004/EMR-1 and No. 9/271(86)/2004/EMR-1, respectively].
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Tyagi.
The nucleotide sequence reported in this paper has been submitted to (database) under accession numbers: Acc. No. EC268400–EC268685; EC365167–EC365455.
An erratum to this article can be found at http://dx.doi.org/10.1007/s00438-009-0441-y
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Govind, G., Vokkaliga ThammeGowda, H., Jayaker Kalaiarasi, P. et al. Identification and functional validation of a unique set of drought induced genes preferentially expressed in response to gradual water stress in peanut. Mol Genet Genomics 281, 591–605 (2009). https://doi.org/10.1007/s00438-009-0432-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-009-0432-z