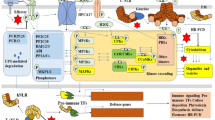
Abstract
Main conclusion
Identification and functional characterization of plant pathogen effectors promise to ameliorate future research and develop effective and sustainable strategies for controlling or containing crop diseases.
Wheat is the second most important food crop of the world after rice. Rust pathogens, one of the major biotic stresses in wheat production, are capable of threatening the world food security. Understanding the molecular basis of plant–pathogen interactions is essential for devising novel strategies for resistance breeding and disease management. Now, it has been established that effectors, the proteins secreted by pathogens, play a key role in plant–pathogen interactions. Therefore, effector biology has emerged as one of the most important research fields in plant biology. Recent advances in genomics and bioinformatics have allowed identification of a large repertoire of candidate effectors, while the evolving high-throughput tools have continued to assist in their functional characterization. The repertoires of effectors have become an important resource for better understanding of effector biology of pathosystems and resistance breeding of crop plants. In recent years, a significant progress has been made in the field of rust effector biology. This review describes the recent advances in effector biology of obligate fungal pathogens, identification and functional analysis of wheat rust pathogens effectors and the potential applications of effectors in molecular plant biology and rust resistance breeding in wheat.



Similar content being viewed by others
References
Ali S, Laurie JD, Linning R, Cervantes-Chavez JA, Gaudet D, Bakkeren G (2014) An immunity-triggering effector from the Barley smut fungus Ustilago hordei resides in an Ustilaginaceae-specific cluster bearing signs of transposable element-assisted evolution. PLoS Pathog 10:e1004223. https://doi.org/10.1371/journal.ppat.1004223
Arazoe T, Miyoshi K, Yamato T, Ogawa T, Ohsato S, Arie T, Kuwata S (2015) Tailor-made CRISPR/Cas system for highly efficient targeted gene replacement in the rice blast fungus. Biotechnol Bioeng 112:2543–2549
Arora S, Steuernagel B et al (2019) Resistance gene cloning from a wild crop relative by sequence capture and association genetics. Nat Biotechnol 37:139–143
Ayliffe M, Devilla R et al (2011a) Nonhost resistance of rice to rust pathogens. Mol Plant Microbe Interact 24(10):1143–1155
Ayliffe M, Jin Y et al (2011b) Determining the basis of nonhost resistance in rice to cereal rusts. Euphytica 179:33–40
Bai Y, Pavan S et al (2008) Naturally occurring broad-spectrum powdery mildew resistance in a Central American tomato accession is caused by loss of Mlo function. Mol Plant Microbe Interact 21(1):30–39
Bai S, Liu J et al (2012) Structure-function analysis of barley NLR immune receptor MLA10 reveals its cell compartment specific activity in cell death and disease resistance. PLoS Pathog 8(6):e1002752
Beck M, Heard W et al (2012) The INs and OUTs of pattern recognition receptors at the cell surface. Curr Opin Plant Biol 15:367–374
Berger S, Benediktyova Z et al (2007) Visualization of dynamics of plant–pathogen interaction by novel combination of chlorophyll fluorescence imaging and statistical analysis: differential effects of virulent and avirulent strains of P. syringae and of oxylipins on A. thaliana. J Exp Bot 58:797–806
Bozkurt TO, Schornack S et al (2011) Phytophthora infestans effector AVRblb2 prevents secretion of a plant immune protease at the haustorial interface. Proc Natl Acad Sci USA 108:20832–20837
Bruce M, Neugebauer KA et al (2014) Using transcription of six Puccinia triticina races to identify the effective secretome during infection of wheat. Front Plant Sci 4:520
Büschges R, Hollricher K et al (1997) The barley Mlo gene: a novel control element of plant pathogen resistance. Cell 88(5):695–705
Bushnell WR, Gay JL (1978) Accumulation of solutes in relation to the structure and function of haustoria in powdery mildews. In: Spencer DM (ed) The powdery mildews. Academic Press, London, pp 183–235
Caillaud MC, Piquerez SJ (2012) Subcellular localization of the HpaRxLR effector repertoire identifies a tonoplast-associated protein HaRxL17 that confers enhanced plant susceptibility. Plant J 69(2):252–265
Caillaud MC, Asai S et al (2013) A downy mildew effector attenuates salicylic acid-triggered immunity in Arabidopsis by interacting with the host mediator complex. PLoS Biol 11:e1001732
Cantu D, Govindarajulu M et al (2011) Next generation sequencing provides rapid access to the genome of Puccinia striiformis f. sp tritici, the causal agent of wheat stripe rust. PLoS One 6:8. https://doi.org/10.1371/journal.pone.0024230
Cantu D, Segovia V, Maclean D, Bayles R, Chen X, Kamoun S et al (2013) Genome analyses of the wheat yellow (stripe) rust pathogen Puccinia striiformis f. sp. tritici reveal polymorphic and haustorial expressed secreted proteins as candidate effectors. BMC Genom 14(1):270
Chaudhari P, Ahmed B et al (2014) Effector biology during biotrophic invasion of plant cells. Virulence 5(7):703–709
Chen XM, Penman L, Wan AM et al (2010) Virulence races of Puccinia striiformis f. sp. tritici in 2006 and 2007 and development of wheat stripe rust and distributions, dynamics, and evolutionary relationships of races from 2000 to 2007 in the United States. Can J Plant Pathol 32:315–333
Chen YE, Cui JM et al (2015) Influence of stripe rust infection on the photosynthetic characteristics and antioxidant system of susceptible and resistant wheat cultivars at the adult plant stage. Front Plant Sci 6:779. https://doi.org/10.3389/fpls.2015.00779
Chen J, Upadhyaya NM et al (2017) Loss of AvrSr50 by somatic exchange in stem rust leads to virulence for Sr50 resistance in wheat. Science 358(6370):1607–1610
Chen S, Guo Y et al (2018) Mapping and characterization of wheat stem rust resistance genes SrTm5 and Sr60 from Triticum monococcum. Theor Appl Genet 131(3):625–635
Cheng Y, Yao J et al (2015) Cytological and molecular analysis of nonhost resistance in rice to wheat powdery mildew and leaf rust pathogens. Protoplasma 252:1167–1179. https://doi.org/10.1007/s00709-014-0750-9
Cheng Y, Wu K et al (2017) PST ha5a23, a candidate effector from the obligate biotrophic pathogen Puccinia striiformis f sp tritici, is involved in plant defense suppression and rust pathogenicity. Environ Microbiol 19(5):1717–1729
Clancy MJ, Shambaugh ME et al (2002) Induction of sporulation in Saccharomyces cerevisiae leads to the formation of N6-methyladenosine in mRNA: a potential mechanism for the activity of the IME4 gene. Nucleic Acids Res 30:4509–4518. https://doi.org/10.1093/nar/gkf573
Cloutier S, McCallum BD et al (2007) Leaf rust resistance gene Lr1, isolated from bread wheat (Triticum aestivum L.) is a member of the large psr567 gene family. Plant Mol Biol 65:93–106
Coll NS, Epple P, Dangl JL (2011) Programmed cell death in the plant immune system. Cell Death Differ 18:1247–1256. https://doi.org/10.1038/cdd.2011.37
Cunningham FJ, Goh NS, Demirer GS, Matos JL, Landry MP (2018) Nanoparticle-mediated delivery towards advancing plant genetic engineering. Trends Biotechnol. https://doi.org/10.1016/j.tibtech.2018.03.009
Cuomo CA, Bakkeren G et al (2017) Comparative analysis highlights variable genome content of wheat rusts and divergence of the mating loci. G3 7:361–376. https://doi.org/10.1534/g3.116.032797
Dagvadorj B, Ozketen AC, Andac A, Duggan C, Bozkurt TO, Akkaya MS (2017) A Puccinia striiformis f. sp. tritici secreted protein activates plant immunity at the cell surface. Sci Rep 7:1141. https://doi.org/10.1038/s41598-017-01100-z
Dean RA, Talbot NJ et al (2005) The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434:980–986
Dean R, Van Kan JA et al (2012) The Top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol 13:414–430
Delaunois B, Jeandet P (2014) Uncovering plant–pathogen crosstalk through apoplastic proteomic studies. Front Plant Sci 5:249. https://doi.org/10.3389/fpls.2014.00249
Delventhal R, Rajaraman J et al (2017) A comparative analysis of nonhost resistance across the two Triticeae crop species wheat and barley. BMC Plant Biol 17(1):232
dit Frey NF, Robatzek S (2009) Trafficking vesicles: pro or contra pathogens? Curr Opin Plant Biol 12:437–443
Doehlemann G, Hemetsberger C (2013) Apoplastic immunity and its suppression by filamentous plant pathogens. New Phytol 198:1001–1016. https://doi.org/10.1111/nph.12277
Dou D, Kale SD et al (2008) RXLR-mediated entry of Phytophthora sojae effector Avr1b into soybean cells does not require pathogen-encoded machinery. Plant Cell 20:1930–1947
Dou D, Kale SD, Liu T et al (2010) Different domains of Phytophthora sojae effector Avr4/6 are recognized by soybean resistance genes Rps4 and Rps6. Mol Plant Microbe Interact 23:425–435. https://doi.org/10.1094/mpmi-23-4-0425
Downie RC, Bouvet L, Furuki E, Gosman N, Gardner KA, Mackay IJ, Tan KC (2018) Assessing European wheat sensitivities to Parastagonospora nodorum necrotrophic effectors and fine-mapping the Snn3-B1 locus conferring sensitivity to the effector SnTox3. Front Plant Sci 9:881
Du J, Vleeshouwers VG (2014) The do’s and don’ts of effectoromics. Methods Mol Biol 1127:257–268
Duplessis S, Cuomo CA, Lin YC et al (2011) Obligate biotrophy features unraveled by the genomic analysis of rust fungi. Proc Natl Acad Sci USA 108:9166–9171. https://doi.org/10.1073/pnas.1019315108
Fabro G, Steinbrenner J et al (2011) Multiple candidate effectors from the oomycete pathogen Hyaloperonospora arabidopsidis suppress host plant immunity. PLoS Pathog 7(11):e1002348
Fan J, Doerner P (2012) Genetic and molecular basis of nonhost disease resistance: complex, yes; silver bullet, no. Curr Opin Plant Biol 15(4):400–406
Faris JD, Liu Z, Xu SS (2013) Genetics of tan spot resistance in wheat. Theor Appl Genet 126:2197–2217
Ferreira RM, Moreira LM et al (2016) Unravelling potential virulence factor candidates in Xanthomonas citri subsp. citri by secretome analysis. Peer J 4:e1734. https://doi.org/10.7717/peerj.1734
Fetch T, Zegeye T, Park R, Hodson D, Wanyera R (2016) Detection of wheat stem rust races TTHSK and PTKTK in the Ug99 race group in Kenya in 2014. Plant Dis 100:1495
Feuillet C, Travella S, Stein N et al (2003) Map-based isolation of the leaf rust disease resistance gene Lr10 from the hexaploid wheat (Triticum aestivum L.) genome. Proc Natl Acad Sci USA 100(25):15253–15258
Fradin EF, Abd-El-Haliem A, Masini L, van den Berg GCM, Joosten MHAJ, Thomma BPHJ (2011) Interfamily transfer of tomato ve1 mediates Verticillium resistance in Arabidopsis. Plant Physiol 156:2255–2265
Fu D, Uauy C et al (2009) A kinase-START gene confers temperature-dependent resistance to wheat stripe rust. Science 323:1357–1360
Giesbers AK, Pelgrom AJ et al (2017) Effector-mediated discovery of a novel resistance gene against Bremia lactucae in a nonhost lettuce species. New Phytol 216(3):915–926
Gijzen M, Ishmael C, Shrestha SD (2014) Epigenetic control of effectors in plant pathogens. Front Plant Sci 5:638. https://doi.org/10.3389/fpls.2014.00638
Gilroy EM, Breen S et al (2011) Presence/absence, differential expression and sequence polymorphisms between PiAVR2 and PiAVR2-like in Phytophthora infestans determine virulence on R2 plants. New Phytol 191:763–776
Gout L, Fudal I, Kuhn ML, Blaise F, Eckert M, Cattolico L et al (2006) Lost in the middle of nowhere: the AvrLm1 avirulence gene of the Dothideomycete Leptosphaeria maculans. Mol Microbiol 60:67–80
Griffe LL (2017) Applying effectoromics and genomics to identify resistance against Rhynchosporium commune in barley. Doctoral dissertation, University of Dundee
Gust AA, Brunner F, Nürnberger T (2010) Biotechnological concepts for improving plant innate immunity. Curr Opin Biotechnol 21(2):204–210
Haas BJ, Kamoun S, Zody MC, Jiang RHY, Handsaker RE, Cano LM et al (2009) Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature 461:393–398
Hein I, Birch PR et al (2009) Progress in mapping and cloning qualitative and quantitative resistance against Phytophthora infestans in potato and its wild relatives. Potato Res 52(3):215–227
Hogenhout SA, Van der Hoorn RA, Terauchi R, Kamoun S (2009) Emerging concepts in effector biology of plant-associated organisms. Mol Plant Microbe Interact 22(2):115–122
Huang L, Brooks SA, Li W et al (2003) Map-based cloning of leaf rust resistance gene Lr21 from the large and polyploid genome of bread wheat. Genetics 164:655–664
Iyer AS, McCouch SR (2004) The rice bacterial blight resistance gene xa5 encodes a novel form of disease resistance. Mol Plant Microbe Interact 17(12):1348–1354
Jafary H, Albertazzi G et al (2008) High diversity of genes for nonhost resistance of barley to heterologous rust fungi. Genetics 178:2327–2339. https://doi.org/10.1534/genetics.107.077552
Janik K, Stellmach H, Mittelberger C, Hause B (2019) Characterization of phytoplasmal effector protein interaction with proteinaceous plant host targets using bimolecular fluorescence complementation (BiFC). In: Musetti R, Pagliari L (eds) Phytoplasmas. Methods in Molecular Biology, vol 1875. Humana Press, New York
Jia Y, McAdams SA et al (2000) Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J 19(15):4004–4014
Jones JD, Dangl JL (2006) The plant immune system. Nature 444(7117):323
Kamoun S (2007) Groovy times: filamentous pathogen effectors revealed. Curr Opin Plant Biol 10:358–365
Kamper J, Kahmann R et al (2006) Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature 444:97–101
Kawashima CG, Guimaraes GA et al (2016) A pigeonpea gene confers resistance to Asian soybean rust in soybean. Nat Biotechnol 34:661–665
Kay S, Hahn S, Marois E, Hause G, Bonas U (2007) A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science 318(5850):648–651
Kiran K, Rawal HC et al (2016) Draft genome of the wheat rust pathogen (Puccinia triticina) unravels genome-wide structural variations during evolution. Genome Biol Evol 8(9):2702–2721. https://doi.org/10.1093/gbe/evw197
Kiran K, Rawal HC et al (2017) Dissection of genomic features and variations of three pathotypes of Puccinia striiformis through whole genome sequencing. Sci Rep 15:85. https://doi.org/10.1038/srep42419
Klymiuk V, Yaniv E, Huang L (2018) Cloning of the wheat Yr15 resistance gene sheds light on the plant tandem kinase-pseudokinase family. Nat Commun 9:e3735. https://doi.org/10.1038/s41467-018-06138-9
Koh S, André A et al (2005) Arabidopsis thaliana subcellular responses to compatible Erysiphe cichoracearum infections. Plant J 44:516–529. https://doi.org/10.1111/j.1365-313x.2005.02545.x
Krattinger SG, Lagudah ES et al (2009) A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 323:1360–1363
Lebrun MH, Langin T et al. (2016) Wheat effector assisted breeding for resistance to fungal pathogens (WEAB). Presented at Journées Jean Chevaugeon 2016 (JJC)—11èmes Rencontres de Phytopathologie—Mycologie, SociétéFrançaise de Phytopathologie (SFP), Aussois, FRA (2016-01-25—2016-01-29). 49. https://prodinra.inra.fr/record/347885
Lee WS, Hammond-Kosack KE, Kanyuka K (2012) Barley stripe mosaic virus-mediated tools for investigating gene function in cereal plants and their pathogens: VIGS, HIGS, VOX. Plant Physiol 160(2):582–590. https://doi.org/10.1104/pp.112.203489
Lee HA, Kim SY et al (2014) Multiple recognition of RXLR effectors is associated with nonhost resistance of pepper against Phytophthora infestans. New Phytol 203(3):926–938
Lenman M, Ali A et al (2016) Effector-driven marker development and cloning of resistance genes against Phytophthora infestans in potato breeding clone SW93-1015. Theor Appl Genet 129(1):105–115
Leonelli L, Erickson E, Lyska D, Niyogi KK (2016) Transient expression in Nicotiana benthamiana for rapid functional analysis of genes involved in non-photochemical quenching and carotenoid biosynthesis. Plant J 88(3):375–386
Li H, Goodwin PH et al (2012) Microscopy and proteomic analysis of the non-host resistance of Oryza sativa to the wheat leaf rust fungus, Puccinia triticina f. sp. tritici. Plant Cell Rep 31:637–650. https://doi.org/10.1007/s00299-011-1181-0
Lipka U, Fuchs R et al (2010) Live and let die-Arabidopsis nonhost resistance to powdery mildews. Eur J Cell Biol 89:194–199. https://doi.org/10.1016/j.ejcb.2009.11.011
Liu W, Frick M, Huel R et al (2014) The stripe rust resistance gene Yr10 encodes an evolutionary-conserved and unique CC-NBS–LRR sequence in wheat. Mol Plant 7:1740–1755. https://doi.org/10.1093/mp/ssu112
Liu R, Chen L, Jiang Y, Zhou Z, Zou G (2015) Efficient genome editing in filamentous fungus Trichoderma reesei using the CRISPR/Cas9 system. Cell Discov 1:15007
Liu CH, Pedersen C et al (2016) The stripe rust fungal effector PEC6 suppresses pattern-triggered immunity in a host species-independent manner and interacts with adenosine kinases. New Phytol 15:85. https://doi.org/10.1111/nph.14034
Lowe I, Cantu D, Dubcovsky J (2011) Durable resistance to the wheat rusts: integrating systems biology and traditional phenotype-based research methods to guide the deployment of resistance genes. Euphytica 179(1):69–79
Lu YJ, Schornack S, Spallek T et al (2012) Patterns of plant subcellular responses to successful oomycete infections reveal differences in host cell reprogramming and endocytic trafficking. Cell Microbiol 14:682–697
Mackie AJ, Roberts AM et al (1993) Glycoproteins recognized by monoclonal antibodies UB7, UB8, and UB10 are expressed early in the development of pea powdery mildew haustoria. Physiol Mol Plant Pathol 43:135–146. https://doi.org/10.1006/pmpp.1993.1046
Maekawa T, Kracher B et al (2012) Conservation of NLR-triggered immunity across plant lineages. Proc Natl Acad Sci USA 109(49):20119–20123
Mago R, Zhang P et al (2015) The wheat Sr50 gene reveals rich diversity at a cereal disease resistance locus. Nat Plants 1:15186
Marchal C et al (2018) BED-domain-containing immune receptors confer diverse resistance spectra to yellow rust. Nat Plants. https://doi.org/10.1038/s41477-018-0236-4
McDonald MC, Ahren D, Simpfendorfer S, Milgate A, Solomon PS (2018) The discovery of the virulence gene ToxA in the wheat and barley pathogen Bipolaris sorokinian. Mol Plant Pathol 19(2):432–439. https://doi.org/10.1111/mpp.12535
McLellan H, Boevink PC et al (2013) An RxLR effector from Phytophthora infestans prevents re-localisation of two plant NAC transcription factors from the endoplasmic reticulum to the nucleus. PLoS Pathog 9:e1003670
Mengiste T (2012) Plant immunity to necrotrophs. Ann Rev Phytopathol 50:267–294
Miller KE, Kim Y, Huh WK, Park HO (2015) Bimolecular fluorescence complementation (BiFC) analysis: advances and recent applications for genome-wide interaction studies. J Mol Biol 427:2039–2055
Moore JW, Herrera-Foessel S et al (2015) A recently evolved hexose transporter variant confers resistance to multiple pathogens in wheat. Nat Genet 47:1494–1498
Niks RE, Marcel TC (2009) Nonhost and basal resistance: how to explain specificity? New Phytol 182(4):817–828
Nirmala J, Drader T et al (2011) Concerted action of two avirulent spore effectors activates reaction to Puccinia graminis 1 (Rpg1)-mediated cereal stem rust resistance. Proc Natl Acad Sci USA 108(35):14676–14681
Nødvig CS, Nielsen JB, Kogle ME, Mortensen UH (2015) A CRISPR–Cas9 system for genetic engineering of filamentous fungi. PLoS One 10:e0133085
Nowara D, Gay A et al (2010) HIGS: host-induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis. Plant Cell 22:3130–3141. https://doi.org/10.1105/tpc.110.077040
Pais M, Win J, Yoshida K, Etherington GJ, Cano LM, Raffaele S, Banfield MJ, Jones A, Kamoun S, Saunders DGO (2013) From pathogen genomes to host plant processes: the power of plant parasitic oomycetes. Genom Biol 14:211. https://doi.org/10.1186/gb-2013-14-6-211
Panwar V, McCallum B, Bakkeren G (2013a) Host-induced gene silencing of wheat leaf rust fungus Puccinia triticina pathogenicity genes mediated by the Barley stripe mosaic virus. Plant Mol Biol 81(6):595–608
Panwar V, McCallum B, Bakkeren G (2013b) Endogenous silencing of Puccinia triticina pathogenicity genes through in planta-expressed sequences leads to the suppression of rust diseases on wheat. Plant J 73(3):521–532
Panwar V, Jordan M et al (2018) Host-induced silencing of essential genes in Puccinia triticina through transgenic expression of RNAi sequences reduces severity of leaf rust infection in wheat. Plant Biotechnol J 16:1013–1023. https://doi.org/10.1111/pbi.12845
Patkar RN, Naqvi NI (2017) Fungal manipulation of hormone-regulated plant defense. PLoS Pathog 13(6):e1006334
Patkar RN, Benke PI et al (2015) A fungal monooxygenase-derived jasmonate attenuates host innate immunity. Nat Chem Biol 11(9):733–740. https://doi.org/10.1038/nchembio.1885
Patpour M, Hovmøller MS et al (2016) Emergence of virulence to SrTmp in the Ug99 race group of wheat stem rust, Puccinia graminis f. sp. tritici, in Africa. Plant Dis 100:522. https://doi.org/10.1094/pdis-08-15-0938-pdn
Periyannan S, Moore J, Ayliffe M, Bansal U, Wang X, Huang L et al (2013) The gene Sr33, an ortholog of barley Mla genes, encodes resistance to wheat stem rust race Ug99. Science 341:786–788. https://doi.org/10.1126/science.1239028
Petre B, Joly DL, Duplessis S (2014) Effector proteins of rust fungi. Front Plant Sci 5:416
Petre B, Saunders DG et al (2015) Candidate effector proteins of the rust pathogen Melampsora larici-populina target diverse plant cell compartments. Mol Plant Microbe Interact 28:689–700
Petre B, Saunders DG et al (2016) Heterologous expression screens in Nicotiana benthamiana identify a candidate effector of the wheat yellow rust pathogen that associates with processing bodies. PLoS One 11:e0149035
Pohl C, Kiel JA, Driessen AJ, Bovenberg RA, Nygard Y (2016) CRISPR/Cas9 based genome editing of Penicillium chrysogenum. ACS Synth Biol 5:754–764
Prasad P, Bhardwaj SC et al (2016) Ug99: saga, reality and status. Curr Sci 110(9):1614–1616
Qi T, Zhu X et al (2017) Host-induced gene silencing of an important pathogenicity factor PsCPK1 in Puccinia striiformis f. sp. tritici enhances resistance of wheat to stripe rust. Plant Biotechnol J 15:85. https://doi.org/10.1111/pbi.12829
Qiao Y, Liu L et al (2013) Oomycete pathogens encode RNA silencing suppressors. Nat Genet 45:330–333
Rampitsch C, Günel A, Beimcik E, Mauthe W (2015) Proteome of monoclonal antibody-purified haustoria from Puccinia triticina Race-1. Proteomics 15(7):1307–1315
Rigal A, Ma Q, Robert S (2014) Unraveling plant hormone signaling through the use of small molecules. Front Plant Sci 5:373. https://doi.org/10.3389/fpls.2014.00373
Roberts AM, Mackie AJ et al (1993) Molecular differentiation in the extra haustorial membrane of pea powdery mildew haustoria at early and late stages of development. Physiol Mol Plant Pathol 143:147–160. https://doi.org/10.1006/pmpp.1993.1047
Ruud AK, Dieseth JA, Lillemo M (2018) Effects of three Parastagonospora nodorum necrotrophic effectors on spring wheat under Norwegian field conditions. Crop Sci 58:159–168
Saintenac C, Zhang W, Salcedo A, Rouse MN, Trick HN, Akhunov E et al (2013) Identification of wheat gene Sr35 that confers resistance to Ug99 stem rust race group. Science 341:783–786. https://doi.org/10.1126/science.1239022
Salcedo A, Rutter W et al (2017) Variation in the AvrSr35 gene determines Sr35 resistance against wheat stem rust race Ug99. Science 358(6370):1604–1606
Saunders DG, Win J et al (2012) Using hierarchical clustering of secreted protein families to classify and rank candidate effectors of rust fungi. PLoS One 7:e29847. https://doi.org/10.1371/journal.pone.0029847
Savadi S, Prasad P et al (2017) Molecular breeding technologies and strategies for rust resistance in wheat (Triticum aestivum) for sustained food security. Plant Pathol 67:771–791. https://doi.org/10.1111/ppa.12802
Schulze-Lefert P, Panstruga R (2011) A molecular evolutionary concept connecting nonhost resistance, pathogen host range, and pathogen speciation. Trends Plant Sci 16(3):117–125
Schwessinger B, Sperschneider J, Cuddy WS et al (2018) A near-complete haplotype-phased genome of the dikaryotic wheat stripe rust fungus Puccinia striiformis f. sp. tritici reveals high interhaplotype diversity. mBio 9:e02275–e02317. https://doi.org/10.1128/mbio.02275-17
See PT, Iagallo EM, Oliver RP et al (2019) Heterologous expression of the Pyrenophora tritici-repentis effector proteins ToxA and ToxB, and the prevalence of effector sensitivity in Australian cereal crops. Front Microbiol 10:182. https://doi.org/10.3389/fmicb.2019.00182
Segovia V, Bruce M et al (2016) Two small secreted proteins from Puccinia triticina induce reduction of β-glucoronidase transient expression in wheat isolines containing Lr9, Lr24 and Lr26. Can J Plant Pathol 38(1):91–102. https://doi.org/10.1080/07060661.2016.1150884
Selin C, De Kievit TR, Belmonte MF, Fernando WGD (2016) Elucidating the role of effectors in plant-fungal interactions: progress and challenges. Front Microbiol 7:600. https://doi.org/10.3389/fmicb.2016.00600
Shafiei R, Hang C, Kang JG, Loake GJ (2007) Identification of loci controlling non-host disease resistance in Arabidopsis against the leaf rust pathogen Puccinia triticina. Mol Plant Pathol 8:773–784. https://doi.org/10.1111/j.1364-3703.2007.00431.x
Sharma S, Sharma S et al (2013) Deployment of the B urkholderiaglumae type III secretion system as an efficient tool for translocating pathogen effectors to monocot cells. Plant J 74(4):701–712
Shi TQ, Liu GN, Ji RY, Shi K, Song P, Ren LJ, Huang H, Ji XJ (2017) CRISPR/Cas9-based genome editing of the filamentous fungi: the state of the art. Appl Microbiol Biotechnol 101(20):7435–7443. https://doi.org/10.1007/s00253-017-8497-9
Singh RP, Hodson DP et al (2006) Current status, likely migration and strategies to mitigate the threat to wheat production from race Ug99 (TTKS) of stem rust pathogen. CAB Rev 1:054
Sohn KH, Lei R et al (2007) The downy mildew effector proteins ATR1 and ATR13 promote disease susceptibility in Arabidopsis thaliana. Plant Cell 19(12):4077–4090
Sonah H, Deshmukh RK, Bélanger RR (2016) Computational prediction of effector proteins in fungi: opportunities and challenges. Front Plant Sci 7:126. https://doi.org/10.3389/fpls.2016.00126
Song X, Rampitsch C et al (2011) Proteome analysis of wheat leaf rust fungus, Puccinia triticina, infection structures enriched for haustoria. Proteomics 11(5):944–963
Sperschneider J, Williams AH (2015) Evaluation of secretion prediction highlights differing approaches needed for oomycete and fungal effectors. Front Plant Sci 6:1168
Sperschneider J, Gardiner DM et al (2016) EffectorP: predicting fungal effector proteins from secretomes using machine learning. New Phytol 210:743–761
Sperschneider J, Catanzariti AM et al (2017) LOCALIZER: subcellular localization prediction of both plant and effector proteins in the plant cell. Sci Rep 7:44598. https://doi.org/10.1038/srep44598
Stergiopoulos I, de Wit PJ (2009) Fungal effector proteins. Ann Rev Phytopathol 47:233–263
Steuernagel B, Periyannan SK, Hernandez-Pinzon I, Witek K, Rouse MN, Yu G et al (2016) Rapid cloning of disease-resistance genes in plants using mutagenesis and sequence capture. Nat Biotechnol 34:652–655
Tabuchi M, Kawai Y et al (2009) Development of a novel functional high-throughput screening system for pathogen effectors in the yeast Saccharomyces cerevisiae. Biosci Biotechnol Biochem 73(10):2261–2267
Tamura S (1990) Historical aspects of gibberellins. In: Takahashi N, Phinney BO, MacMillan J (eds) Gibberellins. Springer, Berlin, pp 1–8
Tan KC, Phan HT, Rybak K, John E, Chooi YH, Solomon PS, Oliver RP (2015) Functional redundancy of necrotrophic effectors–consequences for exploitation for breeding. Front Plant Sci 6:501
Thind AK, Wicker T et al (2017) Rapid cloning of genes in hexaploid wheat using cultivar-specific long-range chromosome assembly. Nat Biotechnol 35(8):793–796
Thomas WJ, Thireault CA, Kimbrel JA, Chang JH (2009) Recombineering and stable integration of the Pseudomonas syringae pv. syringae 61 hrp/hrc cluster into the genome of the soil bacterium Pseudomonas fluorescens Pf0-1. Plant J 60(5):919–928
Tinoco MLP, Dias BB et al (2010) In vivo trans-specific gene silencing in fungal cells by in planta expression of a double-stranded RNA. BMC Biol 8(1):27
Torto TA, Li S, Styer A, Huitema E, Testa A, Gow NA et al (2003) EST mining and functional expression assays identify extracellular effector proteins from the plant pathogen Phytophthora. Genom Res 13(7):1675–1685
Uhse S, Djamei A (2018) Effectors of plant-colonizing fungi and beyond. PLoS Pathog 14(6):e1006992
Uma B, Rani TS, Podile AR (2011) Warriors at the gate that never sleep: non-host resistance in plants. J Plant Physiol 168(18):2141–2152
Upadhyaya NM, Mago R et al (2014) A bacterial type III secretion assay for delivery of fungal effector proteins into wheat. Mol Plant Microbe Interact 27:255–264. https://doi.org/10.1094/mpmi-07-13-0187-f
Upadhyaya NM, Garnica DP et al (2015) Comparative genomics of Australian isolates of the wheat stem rust pathogen Puccinia graminis f. sp. tritici reveals extensive polymorphism in candidate effector genes. Front Plant Sci 5:759
Vleeshouwers VG, Oliver RP (2014) Effectors as tools in disease resistance breeding against biotrophic, hemibiotrophic, and necrotrophic plant pathogens. Mol Plant Microbe Interact 27(3):196–206
Vleeshouwers VG, Rietman H, Krenek P et al (2008) Effector genomics accelerates discovery and functional profiling of potato disease resistance and Phytophthora infestans avirulence genes. PLoS One 3:e2875
Vleeshouwers VG, Raffaele S et al (2011) Understanding and exploiting late blight resistance in the age of effectors. Ann Rev Phytopathol 49:507–531
Vogel JP, Raab TK et al (2002) PMR6, a pectatelyase-like gene required for powdery mildew susceptibility in Arabidopsis. Plant Cell 14(9):2095–2106
Walton JD, Avis TJ et al (2009) Effectors, effectors et encore des effectors: the XIV International Congress on Molecular-Plant Microbe Interactions, Quebec. Mol Plant Microbe Interact 22:1479–1483
Wang F, Lin R, Feng J et al (2015) TaNAC1 acts as a negative regulator of stripe rust resistance in wheat, enhances susceptibility to Pseudomonas syringae, and promotes lateral root development in transgenic Arabidopsis thaliana. Front Plant Sci 6:108. https://doi.org/10.3389/fpls.2015.00108
Wellings CR, McIntosh RA, Walker J (1987) Puccinia striiformis f. sp. tritici in eastern Australia—possible means of entry and implications for plant quarantine. Plant Pathol 36:239–241
Wu JQ, Sakthikumar S et al (2017) Comparative genomics integrated with association analysis identifies candidate effector genes corresponding to Lr20 in phenotype-paired Puccinia triticina isolates from Australia. Front Plant Sci 8:148
Wulff BB, Horvath DM, Ward ER (2011) Improving immunity in crops: new tactics in an old game. Curr Opin Plant Biol 14(4):468–476
Xia C, Wang M et al (2017) Secretome characterization and correlation analysis reveal putative pathogenicity mechanisms and identify candidate avirulence genes in the wheat stripe rust fungus Puccinia striiformis f. sp. tritici. Front Microbiol 8:2394
Xia C, Wang M, Yin C et al (2018) Genomic insights into host adaptation between the wheat stripe rust pathogen (Puccinia striiformis f. sp. tritici) and the barley stripe rust pathogen (Puccinia striiformis f. sp. hordei). BMC Genomics 19:664. https://doi.org/10.1186/s12864-018-5041-y
Yin CT, Jurgenson JE, Hulbert SH (2011) Development of a host-induced RNAi system in the wheat stripe rust fungus Puccinia striiformis f. sp. tritici. Mol Plant Microbe Interact 24:554–561
Yin C, Park JJ et al (2014) Characterization of a tryptophan 2-monooxygenase gene from Puccinia graminis f. sp. tritici involved in auxin biosynthesis and rust pathogenicity. Mol Plant Microbe Interact 27(3):227–235
Yin C, Downey SI, Klages-Mundt NL, Ramachandran S, Chen X, Szabo LJ et al (2015) Identification of promising host-induced silencing targets among genes preferentially transcribed in haustoria of Puccinia. BMC Genom 16:1791. https://doi.org/10.1186/s12864-015-1791-y
Zhang GQ, Angeles ER et al (1996) RAPD and RFLP mapping of the bacterial blight resistance gene xa-13 in rice. Theor Appl Genet 93(1–2):65–70
Zhang H, Guo J, Voegele RT, Zhang J, Duan Y, Luo H, Kang Z (2012) Functional characterization of calcineurin homologs PsCNA1/PsCNB1 in Puccinia striiformis f. sp. tritici using a host-induced RNAi system. PLoS One 7(11):e49262
Zhang W, Chen S et al (2017) Identification and characterization of Sr13, a tetraploid wheat gene that confers resistance to the Ug99 stem rust race group. Proc Natl Acad Sci USA 114:e9483–e9492
Zhao B, Lin X et al (2005) A maize resistance gene functions against bacterial streak disease in rice. Proc Natl Acad Sci USA 102:15383–15388. https://doi.org/10.1073/pnas.0503023102
Zhao M, Wang J et al (2018) Candidate effector Pst_8713 impairs the plant immunity and contributes to virulence of Puccinia striiformis f. sp. tritici. Front Plant Sci 15:85. https://doi.org/10.3389/fpls.2018.01294
Zheng W, Huang L et al (2013a) High genome heterozygosity and endemic genetic recombination in the wheat stripe rust fungus. Nat Commun 4:2673. https://doi.org/10.1038/ncomms3673
Zheng Z, Nonomura T et al (2013b) Loss of function in Mlo orthologs reduces susceptibility of pepper and tomato to powdery mildew disease caused by Leveillula taurica. PLoS One 8(7):e70723
Zhu X, Qi T et al (2017) Host-induced gene silencing of the MAPKK gene PsFUZ7 confers stable resistance to wheat stripe rust. Plant Physiol 175:1853–1863. https://doi.org/10.1104/pp.17.01223
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Prasad, P., Savadi, S., Bhardwaj, S.C. et al. Rust pathogen effectors: perspectives in resistance breeding. Planta 250, 1–22 (2019). https://doi.org/10.1007/s00425-019-03167-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-019-03167-6