Abstract
Recent pathogenomic research on plant parasitic oomycete effector function and plant host responses has resulted in major conceptual advances in plant pathology, which has been possible thanks to the availability of genome sequences.
Similar content being viewed by others
Introduction
Plants come under attack from a wide diversity of pathogens, of which the oomycetes are a notable example. The oomycetes are a diverse and ancient group of eukaryotic microbes that are classified within the stramenopiles, many of which parasitize plants, and are therefore very important to humans [1]. Plant pathogenic oomycetes target a wide range of plants, resulting in significant damage to agricultural and natural ecosystems. The pathogenic lifestyle of oomycetes can be biotrophic, requiring living plants to survive; necrotrophic, actively killing plant cells and feeding on dead matter; or hemibiotrophic, with an initial biotrophic phase followed by a necrotrophic phase. The most notorious oomycetes belong to the genus Phytophthora ('plant destroyer' in Greek), which, with over 100 species, are arguably the most devastating pathogens of dicotyledonous plants [2]. Phytophthora infestans, the causal agent of potato and tomato late blight, has been a recurrent threat to world agriculture ever since triggering the Irish potato famine in the 1840s [3]. P. infestans is remarkable for its ability to rapidly adapt to resistant plants, and also belongs to a lineage that has experienced repeated host-jumps throughout its evolution [3, 4].
The pathogenic and evolutionary success of P. infestans and other plant parasitic oomycetes can be ascribed to their diverse and rapidly evolving effector gene complements [5]. During infection, certain plant genotypes specifically recognize pathogens and mount active defenses to attenuate pathogen progression. Firstly, plant basal defense responses are activated by the perception of pathogen-associated molecular patterns (PAMPs) by pattern recognition receptors at the plant cell surface, resulting in PAMP-triggered immunity [6]. However, pathogen effector proteins can act as virulence factors to suppress these basal defense mechanisms. A second wave of plant host defense acts largely within the cell through the genetically determined recognition of a subset of pathogen effectors, known as avirulence (AVR) proteins. On certain plant genotypes, these AVR effectors can become a liability for the pathogen because they can be detected by immune receptors, encoded by resistance (R) genes, which initiate effective defense reactions. Recognition of AVR effectors can lead to the activation of effector-triggered immunity (ETI) that often results in a rapid localized host cell death, termed the hypersensitive response, thereby limiting disease progression [6]. However, pathogens can also secrete effectors to suppress ETI and promote disease progression. As a result, effector genes are under dynamic, even opposite, selection pressures, depending on the host plant genotype that the pathogen interacts with.
Despite their importance in agricultural systems and the environment, the oomycetes have been relatively poorly studied. Recent work on oomycete effectors has greatly benefited from high-throughput in planta expression assays to link effector genes to biological activities, and from the ability to mine effectors from genome sequences based on conserved host translocation motifs and their presence in well-defined genome compartments. Such approaches could only be implemented because of the availability of genome sequences. As a consequence, recent research on oomycete effector trafficking, function and evolution resulted in major conceptual advances for plant pathology [5, 7, 8].
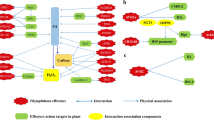
Here, we review how sequencing plant parasitic oomycete genomes and using genomics approaches to study host plant responses to infection has advanced our understanding of oomycete-plant interactions, and ultimately of plant processes that are perturbed by these pathogens. Oomycete pathosystems are an excellent case study illustrating how plant pathogen genomes can be a remarkable resource for basic and applied plant biology (Figure 1).
High-throughput pipeline for using plant pathogen effectors to unravel plant processes. (a) Genome sequences are currently available for a variety of plant pathogens. (b) This allows the computational prediction of candidate effector genes using the knowledge gained from characterized effector proteins. In the case of Phytophthora pathogens, prediction is facilitated by the presence of effector genes in gene-sparse, repeat-rich regions of the genomes, and by the modular structure of effectors. (c) Cloning of these effector genes and (d) expression of candidate effectors in planta using Agrobacterium-mediated transient expression systems such as agroinfiltration (left) and wound inoculation (right). (e) These help us understand diverse plant processes. For instance, (i) identification of host proteins that interact with pathogen effectors can give insights into the plant pathways targeted and perturbed during the infection process. (ii) In addition, effectors can be used as molecular probes to study the structural changes that occur during plant infection at a subcellular level. For instance, effectors can be fused to fluorescent proteins to assess their localization in planta. (iii) The suppression of plant immune responses such as the production of reactive oxygen species (ROS) can also be studied. Blue line shows induction of ROS by flagellin peptide in the absence of effector proteins. This ROS burst is reduced (red) by the expression of an oomycete effector. (iv) The activation of host immunity by effector proteins helps the dissection of plant susceptibility and resistance mechanisms. For example, the hypersensitive response (brown spots on leaves) against effectors transiently expressed in planta can be used to identify immune receptors (R genes) of high value for plant breeding.
Plant parasitic oomycete genome structure
Currently, draft genome sequences are available for ten oomycete species, nine of which are plant pathogens [3, 9–16] (Figure 2). One striking feature of these oomycete genomes is the considerable variability in size, ranging from 37 Mb for the biotrophic plant pathogen Albugo laibachii (an obligate parasite of the model plant Arabidopsis thaliana) , to 240 Mb for the hemibiotrophic P. infestans (which parasitizes tomato and potato). These observed differences in genome size are largely due to the proliferation of transposable elements and repetitive DNA, which in P. infestans account for 74% of the genome content [3]. At 100 Mb, the downy mildew Hyaloperonospora arabidopsidis, an obligate parasite of A. thaliana, also has a relatively large genome size, a recurrent trend in biotrophic oomycete and fungal pathogens [12, 15, 17]. Repeat regions in these expanded genomes tend to be unstable; they may promote genome duplication and shuffling, increased rates of mutagenesis and gene silencing [17, 18].
Features of sequenced oomycete pathogen genomes. The representative phylogeny depicts oomycete pathogens with sequenced genomes and was generated using Interactive Tree Of Life (iTOL) with National Center for Biotechnology Information (NCBI) taxonomy identifiers (branch lengths are arbitrary). Pathogen lifestyles and major variations in effector gene families are indicated along the tree branches. Potential loss of particular effector classes in a lineage is indicated by a red cross. The principal host, genome size, repetitive DNA content (as a percentage of genome size), gene space (the percentage of the genome encoding genes), number of protein-coding genes and number and percentage of proteins encoding predicted secreted proteins (secretome) are indicated from left to right for each pathogen. CRN, Crinkler; ND, not determined.
The genomes of Phytophthora species have a peculiar bipartite architecture. They show a characteristic structure comprising blocks of conserved gene order, containing approximately 90% of core ortholog genes, separated by regions in which gene order is not conserved [3, 11]. Within conserved blocks, gene density is high and repeat and transposable element content is low (Figure 3a). The trend is most extreme in the genome of P. infestans in which the gene-dense and gene-sparse regions can be easily distinguished at the whole-genome level using flanking intergenic region length data (Figure 3b). Remarkably, effector genes mostly populate the gene-sparse, repeat-rich regions of Phytophthora genomes [3, 11] (Figure 3). Because these regions evolve more rapidly than the gene-dense compartments of the genome, P. infestans has been described as having a 'two-speed' genome [19]. The presence of effector genes in plastic genomic regions is thought to promote rapid adaptation to new hosts and evasion of recognition by host immune receptors. It is reminiscent of the occurrence of plant immune receptors (R genes) in rapidly evolving gene clusters of plant genomes [20, 21].
Phytophthora gene-sparse regions are highly enriched in effector genes. (a) Alignment of Phytophthora genomes showing the localization of two avirulence effector genes in gene-sparse regions. The two examples are Phytophthora infestans PiAvr4 and Phytophthora sojae PsAvr4 and PsAvr6. (b) Distribution of Phytophthora infestans RXLR effectors, CRN effectors, and ribosomal protein genes (spots) according to the length of their 5' and 3' flanking intergenic regions. Percentages show the proportion of the gene group falling in the gene-dense region (GDR) and gene-sparse region quadrants as delimited by the 1.5 kb flanking intergenic region length cutoff (dashed lines).
Not all plant pathogenic oomycetes have highly expanded, repeat-rich genomes. Phytophthora ultimum has the lowest level of repetitive DNA at 7%; this may be attributed to the presence of DNA methylases, which have been shown to inhibit repeat expansion and are absent in the P. infestans genome [13]. The relatively compact genome sizes of Albugo candida and A. laibachii (45 and 37 Mb, respectively) may reflect the loss of biosynthetic pathways in these obligate parasites [14, 15]. However, to fully understand genome evolution in parasitic oomycetes, we still need to compare the genomes of the parasites to their saprophytic kin. Fortunately, genome sequencing projects of several non-parasitic oomycete species are in progress.
Effectors and their functions in plants
Effector proteins belong to two classes that target distinct sites in the host plant: apoplastic effectors are secreted into the plant extracellular space, whereas cytoplasmic effectors are translocated inside the plant cell, where they target different subcellular compartments [22]. Both classes of effectors are modular proteins with cleavable amino-terminal secretion signals. Cytoplasmic effectors carry an additional domain after the signal peptide that mediates translocation inside host cells and is defined by conserved motifs, such as the RXLR amino acid sequence [22].
Several functional classes of apoplastic effectors have been described, including enzyme inhibitors and the NEP1-like toxin proteins (NLPs) [22, 23]. One important function of apoplastic effectors is to disable extracellular plant defenses and enable the pathogen to adapt to the protease-rich environment of the plant apoplast [24, 25]. Protease inhibitor effectors that target both plant serine and cysteine proteases have been reported in several oomycetes [3, 11–13]. The P. infestans cystatin-like EPIC1 and EPIC2B inhibit the cysteine proteases PIP1, RCR3 and C14 from tomato and potato host plants [24–26]. Interestingly, C14 is also targeted by the cytoplasmic effector AVRblb2, which interferes with its secretion into the apoplast [27]. Overall, pathogen effectors have proved to be useful probes to identify plant proteases that have roles in immunity. For example, the tomato protease RCR3 is targeted by effectors from a fungus, an oomycete and a nematode, suggesting that it may have an important role in plant apoplastic defenses [24, 28].
There are three validated classes (RXLR, Crinkler (CRN) and CHXC) and one putative class (YxSL[RK]) of cytoplasmic effectors in oomycetes. The best-studied are the RXLR proteins, which include effectors with an avirulence activity, which are detected by plant immune receptors [29]. The carboxy-terminal regions of RXLR proteins carry the biochemical effector activity and about half of these proteins have a similar overall conformation, termed the WY-fold [30–33]. This fold may be an adaptive structural unit that can support effector diversification to gain new functions and/or evade plant host immunity [30, 34]. Among RXLR-WY effectors, AVR3a associates with the potato E3 ubiquitin ligase CMPG1 to suppress the cell death induced by another P. infestans secreted protein, INF1 elicitin [35]. Most RXLR effectors are not predicted to have an enzymatic activity. One exception is Avr3b from Phytophthora sojae, an NADPH and ADP-ribose pyrophosphorylase with a Nudix hydrolase motif that suppresses plant immunity [36]. Although the mechanism by which Avr3b interferes with immunity remains to be determined, Avr3b may have evolved to mimic plant Nudix hydrolases, which are known to act as negative regulators of plant immunity [36].
Effector gene expression
Gene expression profiles in oomycetes exhibit dynamic changes throughout the different developmental stages of their life cycle [37, 38], and dramatic changes occur during infection of host plants [3, 37, 39]. Therefore, high-throughput gene expression analysis by sequencing of cDNA libraries, microarray and RNA-seq technologies are valuable tools to identify candidate virulence factors and to investigate the role of different gene families in the interaction between oomycetes and their hosts [4, 39–42]. Little is known about the molecular clues that trigger these substantial shifts in gene expression and how they are regulated, but recent studies combining genome-wide analysis of promoter sequences with transcriptomics data have enabled the systematic prediction of cis-regulatory motifs that might have a role in stage-specific gene expression [43, 44].
Monitoring gene expression during potato infection showed that about 2.7% of P. infestans genes are induced at least two-fold during infection compared with vegetative growth [3]. In addition, patterns of gene induction correlate with the progression from biotrophic to necrotrophic growth in the host plant (Figure 4a). In total, 79 of the 563 RXLR effector genes predicted in P. infestans display a peak of expression during the biotrophic phase (2 to 3 days post-inoculation, dpi), decaying to basal levels at the necrotrophic phase of infection (4 to 5 dpi) [3]. Comparable induction profiles during biotrophy have been described for P. sojae RXLR effector genes [45]. These expression patterns support the hypothesis that RXLR effectors function mainly during the biotrophic phase of infection. Likewise, several apoplastic effector genes encoding protease inhibitors and cysteine-rich secreted proteins show expression profiles similar to those of RXLR genes in P. infestans [3, 46] (Figure 4a). Finally, a recent analysis of the Phytophthora capsici CRN gene family revealed two classes of CRN effectors with distinct expression patterns during tomato infection [47] (Figure 4b).
Expression patterns of oomycete effector genes. (a) Expression profiles of two representative Phytophthora infestans effector genes in different developmental (mycelia, sporangia and zoospores) and infection (biotrophic and necrotrophic) phases. The life cycle of P. infestans begins with sporangia that are released and travel by wind to new host plants. Under suitable environmental conditions zoospores are released from the sporangium, germinate and penetrate the host plant to initiate infection. As a hemibiotroph, P. infestans undergoes a two-stage infection process, with an initial biotrophic phase followed by a later necrotrophic phase. The data correspond to P. infestans reference strain T30-4 and are based on [3], with expression during infection phases measured in potato. Gene induction values were normalized against the mycelia sample. epi1 encodes an apoplastic effector that functions as protease inhibitor [73] and Avr3a encodes a cytoplasmic RXLR effector [74]. Ubiquitin and pisp3 [75] are included as controls. dpi, days post-inoculation. (b) Crinkler (CRN) gene expression profiles during infection (reproduced from [47]). The heat map shows the expression pattern of full-length Phytophthora capsici CRN genes following tomato infection. Green is downregulated and red is upregulated compared with the median of each sample. Samples were collected at 0, 8, 16, 24, 48 and 72 hours post-infection. Gene classes with distinct expression profiles are indicated on the right.
Little is known about the molecular mechanisms that underpin the transition from biotrophy to necrotrophy, but the P. infestans gene PiNPP1.1, encoding a necrosis-inducing protein, is upregulated during the transition from biotrophic to necrotrophic growth of P. infestans in tomato [48]. SNE1, a gene encoding a suppressor of the host cell death induced by PiNPP1.1, is down-regulated at the same time points of infection [49, 50]. In a similar way, the expression of the P. sojae PsojNIP gene, which encodes a necrosis-inducing protein, is highly induced at late infection stages [51, 52] and it has been suggested that these necrosis-inducing toxins may favor colonization of host plants during the necrotrophic growth [23, 53].
Gain and loss of effector gene induction can also influence the outcome of the interaction with the host plant. Cooke et al. [54] performed infection time course experiments by hybridizing NimbleGen microarrays with cDNA samples from potato leaves inoculated with three different P. infestans strains. This allowed the identification of distinct isolate-specific sets of RXLR genes induced during potato infection. Transcriptional silencing of effector genes is one of the mechanisms that plant pathogenic oomycetes deploy to avoid the activation of host R-gene-mediated immunity. A few examples have been described for races of P. infestans and P. sojae in which the expression of specific RXLR effectors is correlated with their virulence or avirulence on specific host genotypes [29, 55–59]. In P. sojae, these transcriptional polymorphisms have been attributed to insertions or deletions in the promoter region [55, 57, 59].
Host plant responses to effectors
Plants respond to pathogen effectors in various ways depending on the genotypes of both the plant and the invading pathogen. In the first instance when the effector is recognized by pattern recognition receptors, plants can initiate processes associated with PAMP-triggered immunity, such as oxidative bursts, callose deposition and expression of pathogenesis-related genes. Effector proteins can, conversely, act to suppress these processes, promoting pathogen progression and manipulating plant physiological processes. In resistant plant genotypes, products of plant immune receptors, encoded by R genes, may recognize some of these effector proteins and mount ETI-associated processes to restrict pathogen colonization. A major challenge for the plant-pathogen research community is to link effector sequences to plant phenotypes and processes. Functional high-throughput screens of oomycete effector candidates can be performed by in planta transient expression [45, 60, 61] or by delivery of candidate effector proteins by the bacterial type III secretion system [62, 63]. These strategies can give valuable insights into the virulence activities of effector proteins, particularly regarding the suppression of host plant immunity [45, 60, 63]. However, some of these studies need to be analyzed with caution, as discussed by Bozkurt et al. [5]. One example is the screening for suppression of immunity using the mammalian cell-death-inducing protein BAX with a heterologous expression system, which gave high frequencies of cell death suppression when testing P. sojae RXLR effectors [45]. This is because the BAX-induced cell death can be readily suppressed following activation of the unfolded protein response [5]. Another example is the screening of H. arabidopsidis RXLR effectors for the suppression of callose deposition, a component of PAMP-triggered immunity. Given that the frequency of effectors suppressing callose deposition found in this study was high (35 out of 62) [63], follow-up experiments are essential to determine whether this does truly represent identical and redundant defense suppression activities for most of the H. arabidopsidis effectors tested.
Heterologous transient expression of fluorescently tagged effectors in the model plant Nicotiana benthamiana is a useful tool for subcellular localization screens. Caillaud et al. [64] used this approach to test the localization of 49 RXLR effectors of H. arabidopsidis, showing that 33% of the tested proteins localize to the nucleus, another 33% accumulate in the nucleus and cytoplasm and the remaining effectors mainly target various plant cell membranes. In a similar way, Stam et al. [47] analyzed the subcellular localization of 11 diverse P. capsici CRN carboxy-terminal domains and found that all the tested domains target the host nucleus. These findings suggest that the plant cell nucleus has a crucial role in virulence and immunity, and highlights how effectors can help to elucidate the plant cell processes that take place during the interaction with oomycete pathogens.
High-throughput approaches can also be used to study the avirulence activity of effectors and can help to identify and assign functions to new immune receptors [60–62]. Vleeshouwers et al. [61] used a library of RXLR effector genes predicted computationally from the P. infestans genome to screen wild Solanum species (related to potato and tomato) for induction of hypersensitive responses indicating the presence of a plant R gene. A set of 54 effectors were expressed in planta using a Potato virus × (PVX) agroinfection assay optimized for Solanum. This led to the identification of AVRblb1, the effector protein recognized by the Solanum bulbocastanum (wild potato species) resistance protein Rpi-blb1. In a similar study, the S. bulbocastanum resistance gene Rpi-blb2 was co-expressed with a PVX-based library of 62 P. infestans RXLR effector clones in N. benthamiana. This approach allowed the identification of the corresponding effector Avrblb2 [60]. Both Rpi-blb1 and Rpi-blb2 are considered broad-spectrum resistance genes, and the availability of the corresponding Avr genes could assist the use of these R genes in agriculture [29].
In addition, the use of effectors in large-scale screens of germplasm has facilitated the discovery of new resistance genes and their classification into discrete recognition specificities, accelerating the cloning of R genes while avoiding redundant cloning efforts [61, 62, 65]. Effectors can be used to identify R gene homologs in plant species that are more compatible for breeding. These strategies are nicely illustrated by the work of Vleeshouwers et al. [61], in which the screen of several wild Solanum species with a set of predicted P. infestans RXLR effectors led to the discovery and rapid cloning of Solanum stoloniferum Rpi-sto1 and Solanum papita Rpi-pta1, both functionally equivalent to S. bulbocastanum Rpi-blb1, with the additional advantage that S. stoloniferum and S. papita are sexually more compatible with potato, which would facilitate the introgression of this resistance specificity into commercial potato cultivars.
Recently, high-throughput effector screens have also proved useful for dissecting the complex genetic basis of the late blight resistance in the potato cultivar Sarpo Mira, which has both qualitative and quantitative ('field' resistance) components [66]. This analysis identified Rpi-Smira2, an R protein conferring partial resistance on recognition of the P. infestans RXLR effector AVRSmira2. AvrSmira2 is diagnostic of field resistance to late blight and can be used to accelerate the breeding and cloning of Rpi-Smira2, showing that effectors can be used to map linkage of quantitative traits, facilitating quantitative resistance breeding.
Another approach in which knowledge of effectors can assist the deployment of disease resistance against oomycetes is through the monitoring of effector allele diversity in pathogen populations [29, 67, 68]. This can provide valuable information to assess the potential of a given R gene regarding its spectrum and durability, and to design control strategies based on the dynamic distribution of virulence alleles in a given population, allowing the early detection of races that can overcome the deployed R genes. For example, the P. infestans avirulence gene Avrblb1 belongs to the highly ipiO RXLR effector family. The set of ipiO variants present in a given isolate determines the outcome of the interaction between this isolate and host plants carrying the Rpi-blb1 gene, with some combinations conferring avirulence and others overcoming this resistance gene [69, 70]. Therefore, monitoring ipiO diversity in P. infestans populations can tell us whether races are evolving in the field to overcome Rpi-blb1. In addition, genome and transcriptome analysis can be used to determine the set of effector genes present and expressed during infection by isolates of a given genotype, providing information on the R genes that can be deployed to control that particular genotype. This was illustrated by Cooke et al. [54], who showed that an isolate of the P. infestans 13_A2 genotype carries intact coding sequences of Avrblb1, Avrblb2 and Avrvnt1, and that these avirulence genes are induced during infection, suggesting that the cognate R genes (Rpi-blb1, Rpi-blb2 and Rpi-vnt1) could be used to control this aggressive genotype that is predominant in UK fields.
Finally, it is also possible to expand the effector recognition specificity of a given R gene to new virulent alleles by performing artificial evolution by random mutagenesis [29], an approach that has been previously successful when applied to the PVX resistance gene Rx [71].
Conclusions
Pathogenomics has had a substantial impact on both basic and applied plant biology. Effectors have turned out to be effective molecular probes for a variety of plant processes, and knowledge gained from studying effectors has already had an impact on our understanding of basic plant processes, such as immune processes and cellular dynamics [5, 7]. So far, research has focused on a handful of species (Figure 2). In the future, the investigation of effectors from a diverse range of oomycetes, particularly species that alter plant development (such as those causing the deformity called 'witches' brooms') or rely on vertical transmission via plant seeds, is likely to reveal effectors that act beyond plant immunity [72]. Oomycete pathogenomics has also had a great impact on approaches to plant disease resistance breeding, an activity that has been traditionally somewhat 'blind' to the pathogen [29]. Effectors have turned out to be useful tools for probing plant germplasm for new resistance traits and are poised to improve the breeding and deployment of disease resistance genes in agriculture.
References
Lamour K, Kamoun S: Oomycete Genetics and Genomics: Diversity, Interactions, and Research Tools. 2009, New Jersey: John Wiley & Sons
Kroon LP, Brouwer H, de Cock AW, Govers F: The genus Phytophthora anno 2012. Phytopathology. 2012, 102: 348-364. 10.1094/PHYTO-01-11-0025.
Haas BJ, Kamoun S, Zody MC, Jiang RH, Handsaker RE, Cano LM, Grabherr M, Kodira CD, Raffaele S, Torto-Alalibo T, Bozkurt TO, Ah-Fong AM, Alvarado L, Anderson VL, Armstrong MR, Avrova A, Baxter L, Beynon J, Boevink PC, Bollmann SR, Bos JI, Bulone V, Cai G, Cakir C, Carrington JC, Chawner M, Conti L, Costanzo S, Ewan R, Fahlgren N, et al: Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature. 2009, 461: 393-398. 10.1038/nature08358.
Raffaele S, Win J, Cano LM, Kamoun S: Analyses of genome architecture and gene expression reveal novel candidate virulence factors in the secretome of Phytophthora infestans. BMC Genomics. 2010, 11: 637-10.1186/1471-2164-11-637.
Bozkurt TO, Schornack S, Banfield MJ, Kamoun S: Oomycetes, effectors, and all that jazz. Curr Opin Plant Biol. 2012, 15: 483-492. 10.1016/j.pbi.2012.03.008.
Dodds PN, Rathjen JP: Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet. 2010, 11: 539-548.
Stassen JH, Van den Ackerveken G: How do oomycete effectors interfere with plant life?. Curr Opin Plant Biol. 2011, 14: 407-414. 10.1016/j.pbi.2011.05.002.
Win J, Chaparro-Garcia A, Belhaj K, Saunders DG, Yoshida K, Dong S, Schornack S, Zipfel C, Robatzek S, Hogenhout SA, Kamoun S: Effector biology of plant-associated organisms: concepts and perspectives. Cold Spring Harb Symp Quant Biol. 2012, doi: 10.1101/sqb.2012.77.015933
Phytophthora parasitica INRA-310 Sequencing Project, Broad Institute of Harvard and MIT. [http://www.broadinstitute.org/annotation/genome/Phytophthora_parasitica]
Lamour KH, Mudge J, Gobena D, Hurtado-Gonzales OP, Schmutz J, Kuo A, Miller NA, Rice BJ, Raffaele S, Cano LM, Bharti AK, Donahoo RS, Finley S, Huitema E, Hulvey J, Platt D, Salamov A, Savidor A, Sharma R, Stam R, Storey D, Thines M, Win J, Haas BJ, Dinwiddie DL, Jenkins J, Knight JR, Affourtit JP, Han CS, Chertkov O, et al: Genome sequencing and mapping reveal loss of heterozygosity as a mechanism for rapid adaptation in the vegetable pathogen Phytophthora capsici. Mol Plant Microbe Interact. 2012, 25: 1350-1360. 10.1094/MPMI-02-12-0028-R.
Tyler BM, Tripathy S, Zhang X, Dehal P, Jiang RH, Aerts A, Arredondo FD, Baxter L, Bensasson D, Beynon JL, Chapman J, Damasceno CM, Dorrance AE, Dou D, Dickerman AW, Dubchak IL, Garbelotto M, Gijzen M, Gordon SG, Govers F, Grunwald NJ, Huang W, Ivors KL, Jones RW, Kamoun S, Krampis K, Lamour KH, Lee MK, McDonald WH, Medina M, et al: Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science. 2006, 313: 1261-1266. 10.1126/science.1128796.
Baxter L, Tripathy S, Ishaque N, Boot N, Cabral A, Kemen E, Thines M, Ah-Fong A, Anderson R, Badejoko W, Bittner-Eddy P, Boore JL, Chibucos MC, Coates M, Dehal P, Delehaunty K, Dong S, Downton P, Dumas B, Fabro G, Fronick C, Fuerstenberg SI, Fulton L, Gaulin E, Govers F, Hughes L, Humphray S, Jiang RH, Judelson H, Kamoun S, et al: Signatures of adaptation to obligate biotrophy in the Hyaloperonospora arabidopsidis genome. Science. 2010, 330: 1549-1551. 10.1126/science.1195203.
Lévesque CA, Brouwer H, Cano L, Hamilton JP, Holt C, Huitema E, Raffaele S, Robideau GP, Thines M, Win J, Zerillo MM, Beakes GW, Boore JL, Busam D, Dumas B, Ferriera S, Fuerstenberg SI, Gachon CM, Gaulin E, Govers F, Grenville-Briggs L, Horner N, Hostetler J, Jiang RH, Johnson J, Krajaejun T, Lin H, Meijer HJ, Moore B, Morris P, et al: Genome sequence of the necrotrophic plant pathogen Pythium ultimum reveals original pathogenicity mechanisms and effector repertoire. Genome Biol. 2010, 11: R73-10.1186/gb-2010-11-7-r73.
Links MG, Holub E, Jiang RH, Sharpe AG, Hegedus D, Beynon E, Sillito D, Clarke WE, Uzuhashi S, Borhan MH: De novo sequence assembly of Albugo candida reveals a small genome relative to other biotrophic oomycetes. BMC Genomics. 2011, 12: 503-10.1186/1471-2164-12-503.
Kemen E, Gardiner A, Schultz-Larsen T, Kemen AC, Balmuth AL, Robert-Seilaniantz A, Bailey K, Holub E, Studholme DJ, Maclean D, Jones JD: Gene gain and loss during evolution of obligate parasitism in the white rust pathogen of Arabidopsis thaliana. PLoS Biol. 2011, 9: e1001094-10.1371/journal.pbio.1001094.
Saprolegnia genome Sequencing Project, Broad Institute of Harvard and MIT. [http://www.broadinstitute.org/annotation/genome/Saprolegnia_parasitica]
Raffaele S, Kamoun S: Genome evolution in filamentous plant pathogens: why bigger can be better. Nat Rev Microbiol. 2012, 10: 417-430.
Shapiro JA, von Sternberg R: Why repetitive DNA is essential to genome function. Biol Rev. 2005, 80: 227-250.
Raffaele S, Farrer RA, Cano LM, Studholme DJ, MacLean D, Thines M, Jiang RH, Zody MC, Kunjeti SG, Donofrio NM, Meyers BC, Nusbaum C, Kamoun S: Genome evolution following host jumps in the Irish potato famine pathogen lineage. Science. 2010, 330: 1540-1543. 10.1126/science.1193070.
Friedman AR, Baker BJ: The evolution of resistance genes in multi-protein plant resistance systems. Curr Opin Genet Dev. 2007, 17: 493-499. 10.1016/j.gde.2007.08.014.
McHale LK, Truco MJ, Kozik A, Wroblewski T, Ochoa OE, Lahre KA, Knapp SJ, Michelmore RW: The genomic architecture of disease resistance in lettuce. Theor Appl Genet. 2009, 118: 565-580. 10.1007/s00122-008-0921-1.
Kamoun S: A catalogue of the effector secretome of plant pathogenic oomycetes. Annu Rev Phytopathol. 2006, 44: 41-60. 10.1146/annurev.phyto.44.070505.143436.
Gijzen M, Nürnberger T: Nep1-like proteins from plant pathogens: Recruitment and diversification of the NPP1 domain across taxa. Phytochemistry. 2006, 67: 1800-1807. 10.1016/j.phytochem.2005.12.008.
Song J, Win J, Tian M, Schornack S, Kaschani F, Ilyas M, van der Hoorn RA, Kamoun S: Apoplastic effectors secreted by two unrelated eukaryotic plant pathogens target the tomato defense protease Rcr3. Proc Natl Acad Sci USA. 2009, 106: 1654-1659. 10.1073/pnas.0809201106.
Tian M, Win J, Song J, van der Hoorn R, van der Knaap E, Kamoun S: A Phytophthora infestans cystatin-like protein targets a novel tomato papain-like apoplastic protease. Plant Physiol. 2007, 143: 364-377.
Kaschani F, Shabab M, Bozkurt T, Shindo T, Schornack S, Gu C, Ilyas M, Win J, Kamoun S, van der Hoorn RA: An effector-targeted protease contributes to defense against Phytophthora infestans and is under diversifying selection in natural hosts. Plant Physiol. 2010, 154: 1794-1804. 10.1104/pp.110.158030.
Bozkurt TO, Schornack S, Win J, Shindo T, Ilyas M, Oliva R, Cano LM, Jones AM, Huitema E, van der Hoorn RA, Kamoun S: Phytophthora infestans effector AVRblb2 prevents secretion of a plant immune protease at the haustorial interface. Proc Natl Acad Sci USA. 2011, 108: 20832-20837. 10.1073/pnas.1112708109.
Lozano-Torres JL, Wilbers RH, Gawronski P, Boshoven JC, Finkers-Tomczak A, Cordewener JH, America AH, Overmars HA, Van't Klooster JW, Baranowski L, Sobczak M, Ilyas M, van der Hoorn RA, Schots A, de Wit PJ, Bakker J, Goverse A, Smant G: Dual disease resistance mediated by the immune receptor Cf-2 in tomato requires a common virulence target of a fungus and a nematode. Proc Natl Acad Sci USA. 2012, 109: 10119-10124. 10.1073/pnas.1202867109.
Vleeshouwers VG, Raffaele S, Vossen JH, Champouret N, Oliva R, Segretin ME, Rietman H, Cano LM, Lokossou A, Kessel G, Pel MA, Kamoun S: Understanding and exploiting late blight resistance in the age of effectors. Annu Rev Phytopathol. 2011, 49: 507-531. 10.1146/annurev-phyto-072910-095326.
Boutemy LS, King SR, Win J, Hughes RK, Clarke TA, Blumenschein TM, Kamoun S, Banfield MJ: Structures of Phytophthora RXLR effector proteins: a conserved but adaptable fold underpins functional diversity. J Biol Chem. 2011, 286: 35834-35842. 10.1074/jbc.M111.262303.
Chou S, Krasileva KV, Holton JM, Steinbrenner AD, Alber T, Staskawicz BJ: Hyaloperonospora arabidopsidis ATR1 effector is a repeat protein with distributed recognition surfaces. Proc Natl Acad Sci USA. 2011, 108: 13323-13328. 10.1073/pnas.1109791108.
Leonelli L, Pelton J, Schoeffler A, Dahlbeck D, Berger J, Wemmer DE, Staskawicz B: Structural elucidation and functional characterization of the Hyaloperonospora arabidopsidis effector protein ATR13. PLoS Pathog. 2011, 7: e1002428-10.1371/journal.ppat.1002428.
Yaeno T, Li H, Chaparro-Garcia A, Schornack S, Koshiba S, Watanabe S, Kigawa T, Kamoun S, Shirasu K: Phosphatidylinositol monophosphate-binding interface in the oomycete RXLR effector AVR3a is required for its stability in host cells to modulate plant immunity. Proc Natl Acad Sci USA. 2011, 108: 14682-14687. 10.1073/pnas.1106002108.
Win J, Krasileva KV, Kamoun S, Shirasu K, Staskawicz BJ, Banfield MJ: Sequence divergent RXLR effectors share a structural fold conserved across plant pathogenic oomycete species. PLoS Pathog. 2012, 8: e1002400-10.1371/journal.ppat.1002400.
Bos JI, Armstrong MR, Gilroy EM, Boevink PC, Hein I, Taylor RM, Zhendong T, Engelhardt S, Vetukuri RR, Harrower B, Dixelius C, Bryan G, Sadanandom A, Whisson SC, Kamoun S, Birch PR: Phytophthora infestans effector AVR3a is essential for virulence and manipulates plant immunity by stabilizing host E3 ligase CMPG1. Proc Natl Acad Sci USA. 2010, 107: 9909-9914. 10.1073/pnas.0914408107.
Dong S, Yin W, Kong G, Yang X, Qutob D, Chen Q, Kale SD, Sui Y, Zhang Z, Dou D, Zheng X, Gijzen M, Tyler BM, Wang Y: Phytophthora sojae avirulence effector Avr3b is a secreted NADH and ADP-ribose pyrophosphorylase that modulates plant immunity. PLoS Pathog. 2011, 7: e1002353-10.1371/journal.ppat.1002353.
Ye W, Wang X, Tao K, Lu Y, Dai T, Dong S, Dou D, Gijzen M, Wang Y: Digital gene expression profiling of the Phytophthora sojae transcriptome. Mol Plant Microbe Interact. 2011, 24: 1530-1539. 10.1094/MPMI-05-11-0106.
Judelson HS, Ah-Fong AM, Aux G, Avrova AO, Bruce C, Cakir C, da Cunha L, Grenville-Briggs L, Latijnhouwers M, Ligterink W, Meijer HJ, Roberts S, Thurber CS, Whisson SC, Birch PR, Govers F, Kamoun S, van West P, Windass J: Gene expression profiling during asexual development of the late blight pathogen Phytophthora infestans reveals a highly dynamic transcriptome. Mol Plant Microbe Interact. 2008, 21: 433-447. 10.1094/MPMI-21-4-0433.
Savory EA, Adhikari BN, Hamilton JP, Vaillancourt B, Buell CR, Day B: mRNA-Seq analysis of the Pseudoperonospora cubensis transcriptome during cucumber (Cucumis sativus L.) infection. PLoS One. 2012, 7: e35796-10.1371/journal.pone.0035796.
Stassen JH, Seidl MF, Vergeer PW, Nijman IJ, Snel B, Cuppen E, Van den Ackerveken G: Effector identification in the lettuce downy mildew Bremia lactucae by massively parallel transcriptome sequencing. Mol Plant Pathol. 2012, 13: 719-731. 10.1111/j.1364-3703.2011.00780.x.
Cabral A, Stassen JH, Seidl MF, Bautor J, Parker JE, Van den Ackerveken G: Identification of Hyaloperonospora arabidopsidis transcript sequences expressed during infection reveals isolate-specific effectors. PLoS One. 2011, 6: e19328-10.1371/journal.pone.0019328.
Gaulin E, Madoui MA, Bottin A, Jacquet C, Mathe C, Couloux A, Wincker P, Dumas B: Transcriptome of Aphanomyces euteiches: new oomycete putative pathogenicity factors and metabolic pathways. PLoS One. 2008, 3: e1723-10.1371/journal.pone.0001723.
Roy S, Kagda M, Judelson HS: Genome-wide prediction and functional validation of promoter motifs regulating gene expression in spore and infection stages of Phytophthora infestans. PLoS Pathog. 2013, 9: e1003182-10.1371/journal.ppat.1003182.
Seidl MF, Wang RP, Van den Ackerveken G, Govers F, Snel B: Bioinformatic inference of specific and general transcription factor binding sites in the plant pathogen Phytophthora infestans. PLoS One. 2012, 7: e51295-10.1371/journal.pone.0051295.
Wang Q, Han C, Ferreira AO, Yu X, Ye W, Tripathy S, Kale SD, Gu B, Sheng Y, Sui Y, Wang X, Zhang Z, Cheng B, Dong S, Shan W, Zheng X, Dou D, Tyler BM, Wang Y: Transcriptional programming and functional interactions within the Phytophthora sojae RXLR effector repertoire. Plant Cell. 2011, 23: 2064-2086. 10.1105/tpc.111.086082.
Liu Z, Bos JI, Armstrong M, Whisson SC, da Cunha L, Torto-Alalibo T, Win J, Avrova AO, Wright F, Birch PR, Kamoun S: Patterns of diversifying selection in the phytotoxin-like scr74 gene family of Phytophthora infestans. Mol Biol Evol. 2005, 22: 659-672.
Stam R, Jupe J, Howden AJ, Morris JA, Boevink PC, Hedley PE, Huitema E: Identification and characterisation of CRN effectors in Phytophthora capsici shows modularity and functional diversity. PLoS One. 2013, 8: e59517-10.1371/journal.pone.0059517.
Kanneganti TD, Huitema E, Cakir C, Kamoun S: Synergistic interactions of the plant cell death pathways induced by Phytophthora infestans Nepl-like protein PiNPP1.1 and INF1 elicitin. Mol Plant Microbe Interact. 2006, 19: 854-863. 10.1094/MPMI-19-0854.
Kelley BS, Lee SJ, Damasceno CM, Chakravarthy S, Kim BD, Martin GB, Rose JK: A secreted effector protein (SNE1) from Phytophthora infestans is a broadly acting suppressor of programmed cell death. Plant J. 2010, 62: 357-366. 10.1111/j.1365-313X.2010.04160.x.
Lee SJ, Rose JK: Mediation of the transition from biotrophy to necrotrophy in hemibiotrophic plant pathogens by secreted effector proteins. Plant Signal Behav. 2010, 5: 769-772. 10.4161/psb.5.6.11778.
Moy P, Qutob D, Chapman BP, Atkinson I, Gijzen M: Patterns of gene expression upon infection of soybean plants by Phytophthora sojae. Mol Plant Microbe Interact. 2004, 17: 1051-1062. 10.1094/MPMI.2004.17.10.1051.
Qutob D, Kamoun S, Gijzen M: Expression of a Phytophthora sojae necrosis-inducing protein occurs during transition from biotrophy to necrotrophy. Plant J. 2002, 32: 361-373. 10.1046/j.1365-313X.2002.01439.x.
Ottmann C, Luberacki B, Küfner I, Koch W, Brunner F, Weyand M, Mattinen L, Pirhonen M, Anderluh G, Seitz HU, Nürnberger T, Oecking C: A common toxin fold mediates microbial attack and plant defense. Proc Natl Acad Sci USA. 2009, 106: 10359-10364. 10.1073/pnas.0902362106.
Cooke DE, Cano LM, Raffaele S, Bain RA, Cooke LR, Etherington GJ, Deahl KL, Farrer RA, Gilroy EM, Goss EM, Grünwald NJ, Hein I, MacLean D, McNicol JW, Randall E, Oliva RF, Pel MA, Shaw DS, Squires JN, Taylor MC, Vleeshouwers VG, Birch PR, Lees AK, Kamoun S: Genome analyses of an aggressive and invasive lineage of the Irish potato famine pathogen. PLoS Pathog. 2012, 8: e1002940-10.1371/journal.ppat.1002940.
Cui L, Yin W, Dong S, Wang Y: Analysis of polymorphism and transcription of the effector gene Avr1b in Phytophthora sojae isolates from China virulent to Rps1b. Mol Plant Pathol. 2012, 13: 114-122. 10.1111/j.1364-3703.2011.00733.x.
Gilroy EM, Breen S, Whisson SC, Squires J, Hein I, Kaczmarek M, Turnbull D, Boevink PC, Lokossou A, Cano LM, Morales J, Avrova AO, Pritchard L, Randall E, Lees A, Govers F, van West P, Kamoun S, Vleeshouwers VG, Cooke DE, Birch PR: Presence/absence, differential expression and sequence polymorphisms between PiAVR2 and PiAVR2-like in Phytophthora infestans determine virulence on R2 plants. New Phytol. 2011, 191: 763-776. 10.1111/j.1469-8137.2011.03736.x.
Dong S, Yu D, Cui L, Qutob D, Tedman-Jones J, Kale SD, Tyler BM, Wang Y, Gijzen M: Sequence variants of the Phytophthora sojae RXLR effector Avr3a/5 are differentially recognized by Rps3a and Rps5 in soybean. PLoS One. 2011, 6: e20172-10.1371/journal.pone.0020172.
Qutob D, Tedman-Jones J, Dong S, Kuflu K, Pham H, Wang Y, Dou D, Kale SD, Arredondo FD, Tyler BM, Gijzen M: Copy number variation and transcriptional polymorphisms of Phytophthora sojae RXLR effector genes Avr1a and Avr3a. PLoS One. 2009, 4: e5066-10.1371/journal.pone.0005066.
Shan W, Cao M, Leung D, Tyler BM: The Avr1b locus of Phytophthora sojae encodes an elicitor and a regulator required for avirulence on soybean plants carrying resistance gene Rps1b. Mol Plant Microbe Interact. 2004, 17: 394-403. 10.1094/MPMI.2004.17.4.394.
Oh SK, Young C, Lee M, Oliva R, Bozkurt TO, Cano LM, Win J, Bos JI, Liu HY, van Damme M, Morgan W, Choi D, Van der Vossen EA, Vleeshouwers VG, Kamoun S: In planta expression screens of Phytophthora infestans RXLR effectors reveal diverse phenotypes, including activation of the Solanum bulbocastanum disease resistance protein Rpi-blb2. Plant Cell. 2009, 21: 2928-2947. 10.1105/tpc.109.068247.
Vleeshouwers VG, Rietman H, Krenek P, Champouret N, Young C, Oh SK, Wang M, Bouwmeester K, Vosman B, Visser RG, Jacobsen E, Govers F, Kamoun S, Van der Vossen EA: Effector genomics accelerates discovery and functional profiling of potato disease resistance and Phytophthora infestans avirulence genes. PLoS One. 2008, 3: e2875-10.1371/journal.pone.0002875.
Goritschnig S, Krasileva KV, Dahlbeck D, Staskawicz BJ: Computational prediction and molecular characterization of an oomycete effector and the cognate Arabidopsis resistance gene. PLoS Genet. 2012, 8: e1002502-10.1371/journal.pgen.1002502.
Fabro G, Steinbrenner J, Coates M, Ishaque N, Baxter L, Studholme DJ, Korner E, Allen RL, Piquerez SJ, Rougon-Cardoso A, Greenshields D, Lei R, Badel JL, Caillaud MC, Sohn KH, Van den Ackerveken G, Parker JE, Beynon J, Jones JD: Multiple candidate effectors from the oomycete pathogen Hyaloperonospora arabidopsidis suppress host plant immunity. PLoS Pathog. 2011, 7: e1002348-10.1371/journal.ppat.1002348.
Caillaud MC, Piquerez SJ, Fabro G, Steinbrenner J, Ishaque N, Beynon J, Jones JD: Subcellular localization of the Hpa RxLR effector repertoire identifies a tonoplast-associated protein HaRxL17 that confers enhanced plant susceptibility. Plant J. 2012, 69: 252-265. 10.1111/j.1365-313X.2011.04787.x.
Lokossou AA, Park TH, van Arkel G, Arens M, Ruyter-Spira C, Morales J, Whisson SC, Birch PR, Visser RG, Jacobsen E, van der Vossen EA: Exploiting knowledge of R/Avr genes to rapidly clone a new LZ-NBS-LRR family of late blight resistance genes from potato linkage group IV. Mol Plant Microbe Interact. 2009, 22: 630-641. 10.1094/MPMI-22-6-0630.
Rietman H, Bijsterbosch G, Cano LM, Lee HR, Vossen JH, Jacobsen E, Visser RG, Kamoun S, Vleeshouwers VG: Qualitative and quantitative late blight resistance in the potato cultivar Sarpo Mira is determined by the perception of five distinct RXLR effectors. Mol Plant Microbe Interact. 2012, 25: 910-919. 10.1094/MPMI-01-12-0010-R.
Birch PR, Boevink PC, Gilroy EM, Hein I, Pritchard L, Whisson SC: Oomycete RXLR effectors: delivery, functional redundancy and durable disease resistance. Curr Opin Plant Biol. 2008, 11: 373-379. 10.1016/j.pbi.2008.04.005.
Bent AF, Mackey D: Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu Rev Phytopathol. 2007, 45: 399-436. 10.1146/annurev.phyto.45.062806.094427.
Halterman DA, Chen Y, Sopee J, Berduo-Sandoval J, Sanchez-Perez A: Competition between Phytophthora infestans effectors leads to increased aggressiveness on plants containing broad-spectrum late blight resistance. PLoS One. 2010, 5: e10536-10.1371/journal.pone.0010536.
Champouret N, Bouwmeester K, Rietman H, van der Lee T, Maliepaard C, Heupink A, van de Vondervoort PJ, Jacobsen E, Visser RG, van der Vossen EA, Govers F, Vleeshouwers VG: Phytophthora infestans isolates lacking class I ipiO variants are virulent on Rpi-blb1 potato. Mol Plant Microbe Interact. 2009, 22: 1535-1545. 10.1094/MPMI-22-12-1535.
Farnham G, Baulcombe DC: Artificial evolution extends the spectrum of viruses that are targeted by a disease-resistance gene from potato. Proc Natl Acad Sci USA. 2006, 103: 18828-18833. 10.1073/pnas.0605777103.
Thines M, Kamoun S: Oomycete-plant coevolution: recent advances and future prospects. Curr Opin Plant Biol. 2010, 13: 427-433. 10.1016/j.pbi.2010.04.001.
Tian M, Huitema E, Da Cunha L, Torto-Alalibo T, Kamoun S: A Kazal-like extracellular serine protease inhibitor from Phytophthora infestans targets the tomato pathogenesis-related protease P69B. J Biol Chem. 2004, 279: 26370-26377. 10.1074/jbc.M400941200.
Armstrong MR, Whisson SC, Pritchard L, Bos JI, Venter E, Avrova AO, Rehmany AP, Bohme U, Brooks K, Cherevach I, Hamlin N, White B, Fraser A, Lord A, Quail MA, Churcher C, Hall N, Berriman M, Huang S, Kamoun S, Beynon JL, Birch PR: An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proc Natl Acad Sci USA. 2005, 102: 7766-7771. 10.1073/pnas.0500113102.
Kim KS, Judelson HS: Sporangium-specific gene expression in the oomycete phytopathogen Phytophthora infestans. Eukaryot Cell. 2003, 2: 1376-1385. 10.1128/EC.2.6.1376-1385.2003.
Acknowledgements
Research in the SK and MJB laboratories relevant to the areas discussed is supported by funding from BBSRC (UK, grants BB/H019820/1, BB/J004553/1, BB/I019557/1 and BB/J004553/1), ERC (proposal 294608), the John Innes Foundation and the Gatsby Charitable Foundation. KY is supported by a JSPS Postdoctoral Fellowships for Research Abroad, SR a Marie Curie CIG grant (contract 334036) and DGOS a Leverhulme Early Career Fellowship.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
About this article
Cite this article
Pais, M., Win, J., Yoshida, K. et al. From pathogen genomes to host plant processes: the power of plant parasitic oomycetes. Genome Biol 14, 211 (2013). https://doi.org/10.1186/gb-2013-14-6-211
Published:
DOI: https://doi.org/10.1186/gb-2013-14-6-211