Abstract
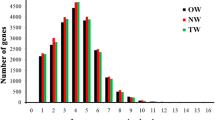
Increasing evidence shows that long non-coding RNAs (lncRNAs) function as important regulatory factors during plant development, but few reports have examined lncRNAs in trees. Here, we report our genome-scale identification and characterization of lncRNAs differentially expressed in the xylem of tension wood, opposite wood and normal wood in Populus tomentosa, by high-throughput RNA sequencing. We identified 1,377 putative lncRNAs by computational analysis, and expression and structure analyses showed that the lncRNAs had lower expression levels and shorter lengths than protein-coding transcripts in Populus. Of the 776 differently expressed (log2FC ≥1 or ≤-1, FDR ≤0.01) lncRNAs, 389 could potentially target 1,151 genes via trans-regulatory effects. Functional annotation of these target genes demonstrated that they are involved in fundamental processes, and in specific mechanisms such as response to stimuli. We also identified 16 target genes involved in wood formation, including cellulose and lignin biosynthesis, suggesting a potential role for lncRNAs in wood formation. In addition, three lncRNAs harbor precursors of four miRNAs, and 25 were potentially targeted by 44 miRNAs where a negative expression relationship between them was detected by qRT-PCR. Thus, a network of interactions among the lncRNAs, miRNAs and mRNAs was constructed, indicating widespread regulatory interactions between non-coding RNAs and mRNAs. Lastly, qRT-PCR validation confirmed the differential expression of these lncRNAs, and revealed that they have tissue-specific expression in P. tomentosa. This study presents the first global identification of lncRNAs and their potential functions in wood formation, providing a starting point for detailed dissection of the functions of lncRNAs in Populus.









Similar content being viewed by others
Abbreviations
- lncRNAs:
-
Long non-coding RNAs
- 4CL:
-
4-Coumarate-CoA ligase
- CCOMT:
-
Caffeoyl-CoA 3-O-methyltransferase
- CESA:
-
Cellulose synthase
- CSLD4:
-
Cellulose synthase-like protein D4
- CW:
-
Compression wood
- GA2ox:
-
Gibberellin 2-oxidase
- GO:
-
Gene ontology
- NW:
-
Normal wood
- OW:
-
Opposite wood
- ppo:
-
Polyphenol oxidase
- TF:
-
Transcription factor
- TW:
-
Tension wood
References
Andersson-Gunnerås S, Mellerowicz EJ, Love J, Segerman B, Ohmiya Y, Coutinho PM, Nilsson P, Henrissat B, Moritz T, Sundberg B (2006) Biosynthesis of cellulose-enriched tension wood in Populus: global analysis of transcripts and metabolites identifies biochemical and developmental regulators in secondary wall biosynthesis. Plant J 45:144–165
Bernal AJ, Yoo C-M, Mutwil M, Jensen JK, Hou G, Blaukopf C, Sørensen I, Blancaflor EB, Scheller HV, Willats WG (2008) Functional analysis of the cellulose synthase-like genes CSLD1, CSLD2, and CSLD4 in tip-growing Arabidopsis cells. Plant Physiol 148:1238–1253
Boerner S, McGinnis KM (2012) Computational identification and functional predictions of long noncoding RNA in Zea mays. PLoS ONE 7:e43047
Bond AM, VanGompel MJ, Sametsky EA, Clark MF, Savage JC, Disterhoft JF, Kohtz JD (2009) Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat Neurosci 12:1020–1027
Cai X, Cullen BR (2007) The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA 13:313–316
Chang S, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11:113–116
Dai X, Zhao PX (2011) psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res 39:W155–W159
Dayan J, Voronin N, Gong F, Sun T-p, Hedden P, Fromm H, Aloni R (2012) Leaf-induced gibberellin signaling is essential for internode elongation, cambial activity, and fiber differentiation in tobacco stems. Plant Cell 24:66–79
Ding J, Lu Q, Ouyang Y, Mao H, Zhang P, Yao J, Xu C, Li X, Xiao J, Zhang Q (2012) A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice. PNAS 109:2654–2659
Du S, Uno H, Yamamoto F (2004) Roles of auxin and gibberellin in gravity-induced tension wood formation in Aesculus turbinata seedlings. IAWA J 25:337–348
Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38:W64–W70
Gibb EA, Brown CJ, Lam WL (2011) The functional role of long non-coding RNA in human carcinomas. Mol Cancer 10:1–17
Goodrich JA, Kugel JF (2006) Non-coding-RNA regulators of RNA polymerase II transcription. Nat Rev Mol Cell Biol 7:612–616
Guo D, Chen F, Inoue K, Blount JW, Dixon RA (2001) Downregulation of caffeic acid 3-O-methyltransferase and caffeoyl CoA 3-O-methyltransferase in transgenic alfalfa: impacts on lignin structure and implications for the biosynthesis of G and S lignin. Plant Cell 13:73–88
Han L, Zhang K, Shi Z, Zhang J, Zhu J, Zhu S, Zhang A, Jia Z, Wang G, Yu S (2012) LncRNA profile of glioblastoma reveals the potential role of lncRNAs in contributing to glioblastoma pathogenesis. Int J Oncol 40:2004–2012
Hu Y, Gai Y, Yin L, Wang X, Feng C, Feng L, Li D, Jiang X-N, Wang D-C (2010) Crystal structures of a Populus tomentosa 4-coumarate: CoA ligase shed light on its enzymatic mechanisms. Plant Cell 22:3093–3104
Israelsson M, Sundberg B, Moritz T (2005) Tissue-specific localization of gibberellins and expression of gibberellin-biosynthetic and signaling genes in wood-forming tissues in aspen. Plant J 44:494–504
Jalali S, Jayaraj GG, Scaria V (2012) Integrative transcriptome analysis suggest processing of a subset of long non-coding RNAs to small RNAs. Biol Direct 7:25
Jalali S, Bhartiya D, Lalwani MK, Sivasubbu S, Scaria V (2013) Systematic transcriptome wide analysis of lncRNA–miRNA interactions. PLoS ONE 8:e53823
Jin J, Zhang H, Kong L, Gao G, Luo J (2013) PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res 42:D1182–D1187
Joseleau J-P, Imai T, Kuroda K, Ruel K (2004) Detection in situ and characterization of lignin in the G-layer of tension wood fibres of Populus deltoides. Planta 219:338–345
Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, Bell I, Cheung E, Drenkow J, Dumais E, Patel S, Helt G, Ganesh M, Ghosh S, Piccolboni A, Sementchenko V, Tammana H, Gingeras TR (2007) RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316:1484–1488
Kwon M (2007) Review: Tension wood as a model system to explore the carbon partitioning between lignin and cellulose biosynthesis in woody plants. J App Biol Chem 50:83–87
Lu S, Sun Y-H, Shi R, Clark C, Li L, Chiang VL (2005) Novel and mechanical stress-responsive microRNAs in Populus trichocarpa that are absent from Arabidopsis. Plant Cell 17:2186–2203
Lu S, Sun YH, Chiang VL (2008) Stress-responsive microRNAs in Populus. Plant J 55:131–151
Lu T, Zhu C, Lu G, Guo Y, Zhou Y, Zhang Z, Zhao Y, Li W, Lu Y, Tang W (2012) Strand-specific RNA-seq reveals widespread occurrence of novel cis-natural antisense transcripts in rice. BMC Genom 13:721
Lu S, Li Q, Wei H, Chang M-J, Tunlaya-Anukit S, Kim H, Liu J, Song J, Sun Y-H, Yuan L (2013) Ptr-miR397a is a negative regulator of laccase genes affecting lignin content in Populus trichocarpa. Proc Natl Acad Sci USA 110:10848–10853
Lukiw WJ, Handley P, Wong L, Crapper McLachlan DR (1992) BC200 RNA in normal human neocortex, non-Alzheimer dementia (NAD), and senile dementia of the Alzheimer type (AD). Neurochem Res 17:591–597
Maury S, Geoffroy P, Legrand M (1999) Tobacco O-methyltransferases involved in phenylpropanoid metabolism. The different caffeoyl-coenzyme a/5-hydroxyferuloyl-coenzyme a 3/5-o-methyltransferase and caffeic acid/5-hydroxyferulic acid 3/5-o-methyltransferase classes have distinct substrate specificities and expression patterns. Plant Physiol 121:215–224
Mellerowicz EJ, Sundberg B (2008) Wood cell walls: biosynthesis, developmental dynamics and their implications for wood properties. Curr Opin Plant Biol 11:293–300
Mercer TR, Dinger ME, Mattick JS (2009) Long non-coding RNAs: insights into functions. Nat Rev Genet 10:155–159
Meyers BC, Axtell MJ, Bartel B, Bartel DP, Baulcombe D, Bowman JL, Cao X, Carrington JC, Chen X, Green PJ (2008) Criteria for annotation of plant MicroRNAs. Plant Cell 20:3186–3190
Nugroho WD, Yamagishi Y, Nakaba S, Fukuhara S, Begum S, Marsoem SN, Ko J-H, Jin H-O, Funada R (2012) Gibberellin is required for the formation of tension wood and stem gravitropism in Acacia mangium seedlings. Ann Bot Lond 110:887–895
Pauli A, Valen E, Lin MF, Garber M, Vastenhouw NL, Levin JZ, Fan L, Sandelin A, Rinn JL, Regev A (2012) Systematic identification of long noncoding RNAs expressed during Zebrafish embryogenesis. Genome Res 22:577–591
Ponting CP, Oliver PL, Reik W (2009) Evolution and functions of long noncoding RNAs. Cell 136:629–641
Puzey JR, Karger A, Axtell M, Kramer EM (2012) Deep annotation of Populus trichocarpa microRNAs from diverse tissue sets. PLoS ONE 7:e33034
Roberts A, Pimentel H, Trapnell C, Pachter L (2011) Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics 27:2325–2329
Saito R, Smoot ME, Ono K, Ruscheinski J, Wang PL, Lotia S, Pico AR, Bader GD, Ideker T (2012) A travel guide to Cytoscape plugins. Nat Methods 9:1069–1076
Schmittgen TD, Lee EJ, Jiang J, Sarkar A, Yang L, Elton TS, Chen C (2008) Real-time PCR quantification of precursor and mature microRNA. Methods 44:31–38
Sigova AA, Mullen AC, Molinie B, Gupta S, Orlando DA, Guenther MG, Almada AE, Lin C, Sharp PA, Giallourakis CC (2013) Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. PNAS 110:2876–2881
Sjödin A, Street NR, Sandberg G, Gustafsson P, Jansson S (2009) The Populus genome integrative explorer (PopGenIE): a new resource for exploring the Populus genome. New Phytol 182:1013–1025
Sun L, Zhang Z, Bailey TL, Perkins AC, Tallack MR, Xu Z, Liu H (2012) Prediction of novel long non-coding RNAs based on RNA-Seq data of mouse Klf1 knockout study. BMC Bioinform 13:331
Tafer H, Hofacker IL (2008) RNAplex: a fast tool for RNA–RNA interaction search. Bioinformatics 24:2657–2663
Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS (2010) Non-coding RNAs: regulators of disease. J Pathol 220:126–139
Timell T (1969) The chemical composition of tension wood. Svensk Papp Tidn 72:173–181
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515
Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7:562–578
Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A et al (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313:1596–1604
Wagner A, Donaldson L, Kim H, Phillips L, Flint H, Steward D, Torr K, Koch G, Schmitt U, Ralph J (2009) Suppression of 4-coumarate-CoA ligase in the coniferous gymnosperm Pinus radiata. Plant Physiol 149:370–383
Wang W, Wang L, Chen C, Xiong G, Tan X-Y, Yang K-Z, Wang Z-C, Zhou Y, Ye D, Chen L-Q (2011a) Arabidopsis CSLD1 and CSLD4 are required for cellulose deposition and normal growth of pollen tubes. J Exp Bot 62:5161–5177
Wang XT, Song XY, Glass CK, Rosenfeld MG (2011b) The long arm of long noncoding RNAs: roles as sensors regulating gene transcriptional programs. CSH Perspect Biol 3:a003756
Yoon JH, Abdelmohsen K, Gorospe M (2013) Post-transcriptional gene regulation by long noncoding RNA. J Mol Biol 425:3723–3730
Yoshida M, Yamamoto H, Okuyama T, Nakamura T (1999) Negative gravitropism and growth stress in GA3-treated branches of Prunus spachiana Kitamura f. spachiana cv. Plenarosea. J Wood Sci 45:368–372
Zhang D, Du Q, Xu B, Zhang Z, Li B (2010) The actin multigene family in Populus: organization, expression and phylogenetic analysis. Mol Genet Genomics 284:105–119
Zhang D, Xu B, Yang X, Zhang Z, Li B (2011) The sucrose synthase gene family in Populus: structure, expression, and evolution. Tree Genet Genomes 7:443–456
Zhang J, Mujahid H, Hou Y, Nallamilli BR, Peng Z (2013a) Plant long ncRNAs: a new frontier for gene regulatory control. Am J Plant Sci 4:1038–1045
Zhang Z, Zhu Z, Watabe K, Zhang X, Bai C, Xu M, Wu F, Mo Y (2013b) Negative regulation of lncRNA GAS5 by miR-21. Cell Death Differ 20:1558–1568
Zhong R, Morrison WH, Himmelsbach DS, Poole FL, Ye Z-H (2000) Essential role of caffeoyl coenzyme A O-methyltransferase in lignin biosynthesis in woody poplar plants. Plant Physiol 124:563–578
Acknowledgments
This work was supported by the State Key Basic Research Program of China (No. 2012CB114506), and the Projects of the National Natural Science Foundation of China (No. 31170622, 30872042), and Shandong Province Agriculture Improved Variety Project (No. 2012213).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chen, J., Quan, M. & Zhang, D. Genome-wide identification of novel long non-coding RNAs in Populus tomentosa tension wood, opposite wood and normal wood xylem by RNA-seq. Planta 241, 125–143 (2015). https://doi.org/10.1007/s00425-014-2168-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-014-2168-1