Abstract
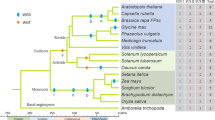
Sucrose synthase is a key enzyme in sucrose metabolism in plant cells, and it is involved in the synthesis of cell wall cellulose. Although the sucrose synthase gene (SUS) family in the model plants Arabidopsis thaliana has been characterized, little is known about this gene family in trees. This study reports the identification of two novel SUS genes in the economically important poplar tree. These genes were expressed predominantly in mature xylem. Using molecular cloning and bioinformatics analysis of the Populus genome, we demonstrated that SUS is a multigene family with seven members that each exhibit distinct but partially overlapping expression patterns. Of particular interest, three SUS genes were preferentially expressed in the stem xylem, suggesting that poplar SUSs are involved in the formation of the secondary cell wall. Gene structural and phylogenetic analyses revealed that the Populus SUS family is composed of four main subgroups that arose before the separation of monocots and dicots. Phylogenetic analyses associated with the tissue- and organ-specific expression patterns. The high intraspecific nucleotide diversity of two SUS genes was detected in the natural population, and the π nonsyn/π syn ratio was significantly less than 1; therefore, SUS genes appear to be evolving in Populus, primarily under purifying selection. This is the first comprehensive study of the SUS gene family in woody plants; the analysis includes genome organization, gene structure, and phylogeny across land plant lineages, as well as expression profiling in Populus.





Similar content being viewed by others
References
Albrecht G, Mustroph A (2003) Localization of sucrose synthase in wheat roots: increased in situ activity of sucrose synthase correlates with cell wall thickening by cellulose deposition under hypoxia. Planta 217:252–260
Barratt DHP, Barber L, Kruger NJ, Smith AM, Wang TL, Martin C (2001) Multiple, distinct isoforms of sucrose synthase in pea. Plant Physiol 127:655–664
Barratt DHP, Derbyshire P, Findlay K, Pike M, Wellner N, Lunn J, Feil R, Simpson C, Maule AJ, Smith AM (2009) Normal growth of Arabidopsis requires cytosolic invertase but not sucrose synthase. Proc Natl Acad Sci USA 106:13124–13129
Baud S, Vaultier MN, Rochat C (2004) Structure and expression profile of the sucrose synthase multigene family in Arabidopsis. J Exp Bot 55:397–409
Bieniawska Z, Barratt DHP, Garlick AP, Thole V, Kruger NJ, Martin C, Zrenner R, Smith AM (2007) Analysis of the sucrose synthase gene family in Arabidopsis. Plant J 49:810–828
Breen AL, Glenn E, Yeager A, Olson MS (2009) Nucleotide diversity among natural populations of a North American poplar (Populus balsamifera, Salicaceae). New Phytol 182:763–773
Chu YG, Su XH, Huang QJ, Zhang XH (2009) Patterns of DNA sequence variation at candidate gene loci in black poplar (Populus nigra L.) as revealed by single nucleotide polymorphisms. Genetica 137:141–150
Clancy M, Hannah LC (2002) Splicing of the maize Sh1 first intron is essential for enhancement of gene expression, and a T-rich motif increases expression without affecting splicing. Plant Physiol 130:918–929
Coleman HD, Yan J, Mansfield SD (2009) Sucrose synthase affects carbon partitioning to increase cellulose production and altered cell wall ultrastructure. Proc Natl Acad Sci USA 106:13118–13123
Demirbas A (2005) Bioethanol from cellulosic materials: a renewable motor fuel from biomass. Energy Sources 27:327–337
Fu YX, Li WH (1993) Statistical tests of neutrality of mutations. Genetics 133:693–709
Fu HY, Kim SY, Park WD (1995) A potato Sus3 sucrose synthase gene contains a context-dependent 3′ element and a leader intron with both positive and negative tissue-specific effects. Plant Cell 7:1395–1403
Haigler CH, Ivanova-Datcheva M, Hogan PS, Salnikov VV, Hwang S, Martin K, Delmer DP (2001) Carbon partitioning to cellulose synthesis. Plant Mol Biol 47:29–51
Hamrick JL, Godt MJW (1996) Effects of life history traits on genetic diversity in plant species. Phil Trans R Soc Lond B 351:1291–1298
Hardin SC, Duncan KA, Huber SC (2006) Determination of structural requirements and probable regulatory effectors for membrane association of maize sucrose synthase 1. Plant Physiol 141:1106–1119
Hauch S, Magel E (1998) Extractable activities and protein content of sucrose phosphate synthase, sucrose synthase and neutral invertase in trunk tissues of Robinia pseudoacacia L. are related to cambial wood production and heart wood formation. Planta 207:266–274
Hirose T, Scofield GN, Terao T (2008) An expression analysis profile for the entire sucrose synthase gene family in rice. Plant Sci 174:534–543
Huang ZH (1992) The study on the climatic regionalization of the distributional region of Populus tomentosa. Journal of Beijing Forestry University 14:26–32
Ingvarsson PK (2005) Nucleotide polymorphism and linkage disequilibrium within and among natural populations of European aspen (Populus tremula L., Salicaceae). Genetics 169:945–953
Ingvarsson PK (2008) Multilocus patterns of nucleotide polymorphism and the demographic history of Populus tremula. Genetics 180:329–340
Joshi CP, Bhandari S, Ranjan P et al (2004) Genomics of cellulose biosynthesis in poplars. New Phytol 164:53–61
Komatsu OA, Moriguchi T, Koyama K, Omura M, Akihama T (2002) Analysis of sucrose synthase genes in citrus suggests different roles and phylogenetic relationships. J Exp Bot 53:61–71
Li X, Weng JK, Chapple C (2008) Improvement of biomass through lignin modification. Plant J 54:569–581
Lunn JE (2002) Evolution of sucrose synthesis. Plant Physiol 128:1490–1500
Luo XY, Xiao YH, Li DM, Luo M, Hou L, Li XB, Yang X, Pei Y (2005) Cloning and analysis of the upstream regulated sequence of a cotton fiber sucrose synthase gene. Chin J Biochem Mol Biol 21:335–340
Mitchell-Olds T, Clauss MJ (2002) Plant evolutionary genomics. Curr Opin Plant Biol 5:74–79
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Ross-Ibarra J, Wright SI, Foxe JP, Kawabe A, DeRose-Wilson L, Gos G, Charlesworth D, Gaut BS (2008) Patterns of polymorphism and demographic history in natural populations of Arabidopsis lyrata. PLoSONE3 6:e2411. doi:10.1371/journal.pone.0002411
Rozas J, Sánchez-Delbarrio JC, Messeguer X, Rozas R (2003) DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496–2497
Ruan YL, Llewellyn DJ, Furbank RT (2003) Suppression of sucrose synthase gene expression represses cotton fiber cell initiation, elongation, and seed development. Plant Cell 15:952–964
Savard L, Li P, Strauss SH, Chase MW, Michaud M, Bousquet J (1994) Chloroplast and nuclear gene sequences indicate late Pennsylvanian time for the last common ancestor of extant seed plants. Proc Natl Acad Sci USA 91:5163–5167
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Tang GQ, Strum A (1999) Antisense repression of sucrose synthase in carrot (Daucus carota L.) affects growth rather than sucrose partitioning. Plant Mol Biol 41:465–479
Tuskan GA, DiFazio S, Jansson S et al (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313:1596–1604
Watterson GA (1975) On the number of segregating sites in genetical models without recombination. Theor Popul Biol 7:188–193
Zhang DQ, Zhang ZY, Yang K (2007) Identification of AFLP markers associated with embryonic root development in Populus tomentosa. Silvae Genet 56:27–32
Zhang DQ, Du QZ, Xu BH, Zhang ZY, Li B (2010a) The actin multigene family in Populus: organization, expression and phylogenetic analysis. Mol Genet Genomics 284:105–119
Zhang DQ, Yang XH, Zhang ZY, Li B (2010b) Expression and nucleotide diversity of the poplar COBL gene. Tree Genet Genomes 6:331–344
Acknowledgments
This work was supported by grants from the Fundamental Research Funds for the Central Universities (No. BLJC200906), the project of National Natural Science Foundation of China (No. 30600479, 30872042), Major Science Foundation of Ministry of Education of China (No. 307006), Foundation for the Author of National Excellent Doctoral Dissertation of PR China (No. 200770), Program for New Century Excellent Talents in University (No. NCET-07-0084), and Introduction of Foreign Advanced Agricultural Science and Technology into China (No. 2009-4-22).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by W. Boerjan
Sequence data from this article have been deposited with the GenBank Data Library under accession numbers GU559727–GU559735 (PtSUS1, PtSUS2, PtrSUS1 to PtrSUS7 cDNA) and HM026865–HM026944 for the population sequences of PtSUS1 and PtSUS2 genes.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
Geographic location of each individual P. tomentosa tree used for nucleotide diversity analysis. The relevant Chinese provinces are shown. Each tree is indicated by a red dot (DOC 1411 kb)
Fig. S2
Nucleotide and deduced amino acid sequences of PtSUS1 (a) and PtSUS2 (b). Numbers on the left refer to the positions of nucleotides or amino acid residues (DOC 52 kb)
Fig. S3
Spatiotemporal expression patterns of PtSUS1 and PtSUS2 in poplar. a Relative transcript levels of PtSUS1 and PtSUS2 in various poplar tissues. b The spatial expression of PtSUS1 and PtSUS2 in the mature xylem of poplar (DOC 29 kb)
Fig. S4
Protein sequence alignment of PtrSUS family members. The numbers on the left are the positions of the amino acids in each protein, with gaps (dashes) included to maximize alignments. Identical and similar amino acids are shaded in pink and blue, respectively (DOC 189 kb)
Table S1
Primers used for sucrose synthase cDNA amplification (DOC 30 kb)
Table S2
Primers used for sucrose synthase gene amplification (DOC 42 kb)
Table S3
List of sucrose synthase gene sequences used in this study (DOC 59 kb)
Table S4
Lengths of gene regions analyzed in this study (DOC 36 kb)
Table S5
The frequencies of nucleotide substitution types in PtSUS1 and PtSUS2 (DOC 35 kb)
Rights and permissions
About this article
Cite this article
Zhang, D., Xu, B., Yang, X. et al. The sucrose synthase gene family in Populus: structure, expression, and evolution. Tree Genetics & Genomes 7, 443–456 (2011). https://doi.org/10.1007/s11295-010-0346-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-010-0346-2