Abstract
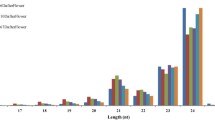
MicroRNAs (miRNAs) are a class of small non-coding RNA molecules that are endogenous regulators of gene expression. miRNAs play a crucial role in cells via degradation of target mRNAs or by inhibition of target protein translation. In the present study, 186 new potentially conserved pear miRNAs belonging to 37 families were identified. The length of mature miRNAs ranged from 19 to 24 nt, and most of the miRNAs (154 out of 186) were 21 nt in length. The length of pre-miRNAs in pear was also found to vary from 62 to 282 nt with an average of 105 ± 43 nt. The potential miRNAs belonged to 29 clusters involving 20 different miRNA families. Using these potential miRNAs, we further scoured of the pear genome and found 326 potential target genes, which included transcription factors, stress responsive genes, and the genes involved in transmembrane transport and signal transduction. Gene ontology analysis of these potential targets suggested that 47 biological processes were potentially regulated by miRNAs, including oxidation–reduction, stress response, transport, etc. KEGG pathway analysis showed that the identified miRNAs were found in 15 metabolism networks which were related to starch and sucrose metabolism, and ascorbate and aldarate metabolism, among others. Our study will help in the further understanding of the essential role of miRNAs in growth and development and stress response of pear.




Similar content being viewed by others
Abbreviations
- GO:
-
Gene ontology
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- miRNAs:
-
MicroRNAs
- MFE:
-
Minimal folding free energy
- MFEI:
-
Minimal folding free energy index
- nt:
-
Nucleotide
- pre-miRNAs:
-
MiRNAs precursor
- NCBI:
-
National center for biotechnology information
- HTGS:
-
High through-put genomic sequences
References
Ambros V (2004) The functions of animal microRNAs. Nature 431:350–355
Ashburner M, Bergman CM (2005) Drosophila melanogaster: a case study of a model genomic sequence and its consequences. Genome Res 15:1661–1667
Aukerman MJ, Sakai H (2003) Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15:2730–2741
Barakat A, Wall P, DiLoreto S, dePamphilis C, Carlson J (2007) Conservation and divergence of microRNAs in populus. BMC Genomics 8:481
Bari R, Datt Pant B, Stitt M, Scheible WR (2006) PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol 141:988–999
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297
Barvkar VT, Pardeshi VC, Kale SM, Qiu SQ, Rollins M, Datla R, Gupta VS, Kadoo NY (2013) Genome-wide identification and characterization of microRNA genes and their targets in flax (Linum usitatissimum). Planta 237:1149–1161
Bonnet E, Wuyts J, Rouzé P, Van de Peer Y (2004) Detection of 91 potential conserved plant microRNAs in Arabidopsis thaliana and Oryza sativa identifies important target genes. P Natl Acad Sci USA 101:11511–11516
Chen X (2004) A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303:2022–2025
Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676
Ding J, Li D, Ohler U, Guan J, Zhou S (2012) Genome-wide search for miRNA-target interactions in Arabidopsis thaliana with an integrated approach. BMC Genomics 13:S3
Dong QH, Han J, Yu HP, Wang C, Zhao MZ, Liu H, Ge AJ, Fang JG (2012) Computational identification of microRNAs in strawberry expressed sequence tags and validation of their precise sequences by miR-RACE. J Hered 103:268–277
Floyd SK, Bowman JL (2004) Gene regulation: ancient microRNA target sequences in plants. Nature 428:485–486
Frazier T, Xie F, Freistaedter A, Burklew C, Zhang B (2010) Identification and characterization of microRNAs and their target genes in tobacco (Nicotiana tabacum). Planta 232:1289–1308
Gandikota M, Birkenbihl RP, Höhmann S, Cardon GH, Saedler H, Huijser P (2007) The miRNA156/157 recognition element in the 3′ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J 49:683–693
Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ (2008) MiRBase: tools for microRNA genomics. Nucleic Acids Res 36:D154–D158
Guo HS, Xie Q, Fei JF, Chua NH (2005) MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell 17:1376–1386
He L, Hannon GJ (2004) MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 5:522–531
Juarez MT, Kui JS, Thomas J, Heller BA, Timmermans MCP (2004) MicroRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428:84–88
Kanehisa M, Goto S (2000) KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28:27–30
Kim JY, Kwak KJ, Jung HJ, Lee HJ, Kang H (2010) MicroRNA402 affects seed germination of Arabidopsis thaliana under stress conditions via targeting DEMETER-LIKE Protein3 mRNA. Plant Cell Physiol 51:1079–1083
Kozomara A, Griffiths-Jones S (2011) MiRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res 39:D152–D157
Leung AKL, Sharp PA (2010) MicroRNA functions in stress responses. Mol Cell 40:205–215
Liu PP, Montgomery TA, Fahlgren N, Kasschau KD, Nonogaki H, Carrington JC (2007) Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J 52:133–146
Liu GQ, Li WS, Zheng PH, Xu T, Chen LJ, Liu DF, Hussain S, Teng YW (2012a) Transcriptomic analysis of ‘Suli’ pear (Pyrus pyrifolia white pear group) buds during the dormancy by RNA-Seq. BMC Genomics 13:700
Liu H, Jin T, Liao RQ, Wan LX, Xu B, Zhou SG, Guan JH (2012b) MiRFANs: an integrated database for Arabidopsis thaliana microRNA function annotations. BMC Plant Biol 12:68
Lu S, Sun YH, Shi R, Clark C, Li L, Chiang VL (2005) Novel and mechanical stress-responsive microRNAs in Populus trichocarpa that are absent from Arabidopsis. Plant Cell 17:2186–2203
Moxon S, Jing R, Szittya G, Schwach F, Rusholme Pilcher RL, Moulton V, Dalmay T (2008) Deep sequencing of tomato short RNAs identifies microRNAs targeting genes involved in fruit ripening. Genome Res 18:1602–1609
Okamuro JK, Caster B, Villarroel R, Van Montagu M, Jofuku KD (1997) The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. P Natl Acad Sci USA 94:7076–7081
Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D (2003) Control of leaf morphogenesis by microRNAs. Nature 425:257–263
Park W, Li J, Song R, Messing J, Chen X (2002) CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol 12:1484–1495
Rubio-Somoza I, Weigel D (2013) Coordination of flower maturation by a regulatory circuit of three microRNAs. PLoS Genet 9:e1003374. doi:10.1371/journal.pgen.1003374
Song C, Jia Q, Fang J, Li F, Wang C, Zhang Z (2010) Computational identification of citrus microRNAs and target analysis in citrus expressed sequence tags. Plant Biol 12:927–934
Sunkar R, Zhu J-K (2004) Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16:2001–2019
Unver T, Budak H (2009) Conserved microRNAs and their targets in model grass species Brachypodium distachyon. Planta 230:659–669
Wang Q, Miyakoda M, Yang W, Khillan J, Stachura DL, Weiss MJ, Nishikura K (2004) Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J Biol Chem 279:4952–4961
Wu J, Wang Z, Shi Z, Zhang S, Ming R, Zhu S, Khan MA, Tao S, Korban SS, Wang H, Chen NJ, Nishio T, Xu X, Cong L, Qi K, Huang X, Wang Y, Zhao X, Wu J, Deng C, Gou C, Zhou W, Yin H, Qin G, Sha Y, Tao Y, Chen H, Yang Y, Song Y, Zhan D, Wang J, Li L, Dai M, Gu C, Wang Y, Shi D, Wang X, Zhang H, Zeng L, Zheng D, Wang C, Chen M, Wang G, Xie L, Sovero V, Sha S, Huang W, Zhang S, Zhang M, Sun J, Xu L, Li Y, Liu X, Li Q, Shen J, Wang J, Paull RE, Bennetzen JL, Wang J, Zhang S (2012) The genome of pear (Pyrus bretschneideri Rehd.). Genome Res 23:396–408
Xia R, Zhu H, An YQ, Beers E, Liu Z (2012) Apple miRNAs and tasiRNAs with novel regulatory networks. Genome Biol 13:R47
Xie FL, Huang SQ, Guo K, Xiang AL, Zhu YY, Nie L, Yang ZM (2007) Computational identification of novel microRNAs and targets in Brassica napus. FEBS Lett 581:1464
Xie FL, Frazier TP, Zhang BH (2010) Identification and characterization of microRNAs and their targets in the bioenergy plant switchgrass (Panicum virgatum). Planta 232:417–434
Xie FL, Frazier TP, Zhang BH (2011) Identification, characterization and expression analysis of microRNAs and their targets in the potato (Solanum tuberosum). Gene 473:8–22
Yin Z, Li C, Han X, Shen F (2008) Identification of conserved microRNAs and their target genes in tomato (Lycopersicon esculentum). Gene 414:60–66
Zhang BH, Pan XP, Anderson TA (2006a) Identification of 188 conserved maize microRNAs and their targets. FEBS Lett 580:3753–3762
Zhang BH, Pan XP, Cannon CH, Cobb GP, Anderson TA (2006b) Conservation and divergence of plant microRNA genes. Plant J 46:243–259
Zhang BH, Pan XP, Cox SB, Cobb GP, Anderson TA (2006c) Evidence that miRNAs are different from other RNAs. Cell Mol Life Sci 63:246–254
Zhang BH, Wang QL, Wang KB, Pan XP, Liu F, Guo TL, Cobb GP, Anderson TA (2007) Identification of cotton microRNAs and their targets. Gene 397:26–37
Zhang BH, Pan XP, Stellwag EJ (2008) Identification of soybean microRNAs and their targets. Planta 229:161–182
Zhang LF, Chia JM, Kumari S, Stein JC, Liu ZJ, Narechania A, Maher CA, Guill K, McMullen MD, Ware D (2009) A genome-wide characterization of microRNA genes in maize. PLoS Genet 5:e1000716. doi:10.1371/journal.pgen.1000716
Zhao B, Ge L, Liang R, Li W, Ruan K, Lin H, Jin Y (2009) Members of miR-169 family are induced by high salinity and transiently inhibit the NF-YA transcription factor. BMC Mol Biol 10:29
Zhou ZS, Huang SQ, Yang ZM (2008) Bioinformatic identification and expression analysis of new microRNAs from Medicago truncatula. Biochem Bioph Res Co 374:538–542
Zhou M, Sun J, Wang QH, Song LQ, Zhao G, Wang HZ, Yang HX, Li X (2011) Genome-wide analysis of clustering patterns and flanking characteristics for plant microRNA genes. FEBS J 278:929–940
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 31272141) and the earmarked fund for Modern Agro-industry Technology Research Systems (nycytx-29-14).
Author information
Authors and Affiliations
Corresponding author
Additional information
Q. Niu and M. Qian contributed equally to this work.
Rights and permissions
About this article
Cite this article
Niu, Q., Qian, M., Liu, G. et al. A genome-wide identification and characterization of mircoRNAs and their targets in ‘Suli’ pear (Pyrus pyrifolia white pear group). Planta 238, 1095–1112 (2013). https://doi.org/10.1007/s00425-013-1954-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-013-1954-5