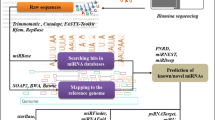
Abstract
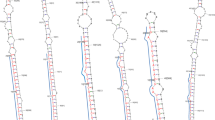
MicroRNAs (miRNAs) are typically non-coding RNAs of 18–26 nucleotides (nts) that are produced endogenously and regulated post-transcriptionally through degradation or translational repression. Since miRNAs are evolutionarily conserved, their preservation is essential for important regulatory functions in plant development, growth, and responses to environmental stress. Sorghum bicolor (sbi) is a valuable food and fodder crop which is grown worldwide. A range of sbi miRNAs were identified so far as being connected to plant development and stress responses. Herein, we employed a variety of bioinformatics tools for miRNA profiling in sbi and a PCR-based platform for the validation of these miRNAs. In total, 74 new conserved sbi miRNAs from 52 miRNA families have been predicted. Using the psRNA Target method, 10613 different protein targets of these predicted miRNAs have been attained. These targets include 54 GO-terms which have substantial targets in the biological, molecular, and cellular processes. We particularly found that the sbi-miR1861c and sbi-miR5050 are involved to regulate sulphur compound biosynthetic process, while the significant spliceosomal complex is regulated by sbi-miR815b and sbi-miR7768b. Also, we report that the pre-ribosome, electron transport chain, cell communication, cellular respiration, protein localization, and photosynthesis are controlled by sbi-miR2907b, sbi-miR530, sbi-miR7749, sbi-miR1858a, sbi-mi7729a, and sbi-miR417, respectively. The identification and validation of these novel sbi miRNAs shall contribute a lot in improving the crop yield and ensure sustainable agriculture.
Graphical Abstract











Similar content being viewed by others
References
Almatroudi, A. (2022). Non-coding RNAs in tuberculosis epidemiology: Platforms and approaches for investigating the Genome’s dark matter. International Journal of Molecular Sciences, 23, 4430.
Md Yusof, K., Rosli, R., Abdullah, M., & Avery-Kiejda, K. A. (2020). The roles of non-coding RNAs in tumor-associated lymph angiogenesis. Cancers, 12, 3290.
Achakzai, H. K., Barozai, M. Y. K., Din, M., Baloch, I. A., & Achakzai, A. K. K. (2018). Identification and annotation of newly conserved microRNAs and their targets in wheat (Triticum aestivum L.). PLoS ONE, 13, e0200033.
Rani, V., & Sengar, R. S. (2022). Biogenesis and mechanisms of microRNA-mediated gene regulation. Biotechnology and Bioengineering, 119, 685–692.
Hajieghrari, B., & Farrokhi, N. (2022). Plant RNA-mediated gene regulatory network. Genomics, 114, 409–442.
Rojas-Pirela, M., Andrade-Alviarez, D., Medina, L., Castillo, C., Liempi, A., Guerrero-Muñoz, J., Ortega, Y., Maya, J. D., Rojas, V., Quiñones, W., & Michels, P. A. (2022). MicroRNAs: Master regulators in host–parasitic protist interactions. Open Biology, 12, 210395.
Kirchner, B. (2022). Functional importance of intra-and extracellular microRNAs and their isoforms in blood and milk (Doctoral dissertation, Technische Universität München).
Barozai, M. Y. K., Irfan, M., Yousaf, R., Ali, I., Qaisar, U., Maqbool, A., Zahoor, M., Rashid, B., Hussnain, T., & Riazuddin, S. (2008). Identification of micro-RNAs in cotton. Plant Physiology and Biochemistry, 46, 739–751.
Din, M., Barozai, M. Y. K., & Baloch, I. A. (2016). Profiling and annotation of microRNAs and their putative target genes in chilli (Capsicum annuum L.) using ESTs. Gene Reports, 5, 62–69.
Xie, F., Frazier, T. P., & Zhang, B. (2010). Identification and characterization of microRNAs and their targets in the bioenergy plant switchgrass (Panicum virgatum). Planta, 232, 417–434.
Barozai, M. Y. K., Ye, Z., Sangireddy, S. R., & Zhou, S. (2018). Bioinformatics profiling and expressional studies of microRNAs in root, stem and leaf of the bioenergy plant switchgrass (Panicum virgatum L.) under drought stress. Agri Gene, 8, 1–8.
Zhang, B., Pan, X., & Stellwag, E. J. (2008). Identification of soybean microRNAs and their targets. Planta, 229, 161–182.
Baloch, I. A., Barozai, M. Y. K., & Din, M. (2018). Bioinformatics prediction and annotation of cherry (Prunus avium L.) microRNAs and their targeted proteins. Turkish Journal of Botany, 42, 382–399.
Din, M., & Barozai, M. Y. K. (2014). Profiling microRNAs and their targets in an important fleshy fruit: Tomato (Solanum lycopersicum). Gene, 535, 198–203.
Barozai, M. Y. K., Baloch, I. A., & Din, M. (2012). Identification of MicroRNAs and their targets in Helianthus. Molecular Biology Reports, 39, 2523–2532.
Barozai, M. Y. K., Qasim, M., Din, M., & Achakzai, A. K. K. (2018). An update on the microRNAs and their targets in unicellular red alga Porphyridium cruentum. Pakistan Journal of Botany, 50, 817–825.
Gul, Z., Barozai, M. Y. K., & Din, M. (2017). In-silico based identification and functional analyses of miRNAs and their targets in Cowpea (Vigna unguiculata L.). Aims Genetics, 4, 138–165.
Baloch, I. A., Barozai, M. Y. K., Din, M., & Achakzai, A. K. K. (2015). Computational identification of 18 microRNAs and their targets in three species of rose. Pakistan Journal of Botany, 47, 1281–1285.
Sohani, S., Chouhan, R., Birla, D., & Patel, L. (2022). Impact of quantities of nitrogen application on infestation of sorghum insect pest. Research Review International Journal of Multidisciplinary, 7, 62–67.
Gyawali, B., Barozai, M. Y. K., & Aziz, A. N. (2021). Comparative expression analysis of microRNAs and their targets in emerging bio-fuel crop sweet sorghum (Sorghum bicolor L.). Plant Gene, 26, 100274.
Katiyar, A., Smita, S., Muthusamy, S. K., Chinnusamy, V., Pandey, D. M., & Bansal, K. C. (2015). Identification of novel drought-responsive microRNAs and trans-acting siRNAs from Sorghum bicolor (L.) Moench by high-throughput sequencing analysis. Frontiers in Plant Science, 6, 506.
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., & Lipman, D. J. (1990). Basic local alignment search tool. Journal of Molecular Biology, 215, 403–410.
Zuker, M. (2003). Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Research, 31, 3406–3415.
Paolacci, A. R., Tanzarella, O. A., Porceddu, E., & Ciaffi, M. (2009). Identification and validation of reference genes for quantitative RT-PCR normalization in wheat. BMC Molecular Biology, 10, 1–27.
Ambros, V., & Lee, R. C. (2004). Identification of microRNAs and other tiny noncoding RNAs by cDNA cloning. Methods in Molecular Biology, 265, 131–158.
Crooks, G. E., Hon, G., Chandonia, J. M., & Brenner, S. E. (2004). WebLogo: A sequence logo generator. Genome research, 14, 1188–1190.
Dai, X., & Zhao, P. X. (2011). psRNATarget: A plant small RNA target analysis server. Nucleic Acids Research, 39, W155–W159.
Tian, T., Liu, Y., Yan, H., You, Q., Yi, X., Du, Z., Xu, W., & Su, Z. (2017). agriGO v2. 0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Research, 45, W122–W129.
Wahid, H. A., Barozai, M. Y. K., & Din, M. (2016). Functional characterization of fifteen hundred transcripts from Ziarat juniper (Juniperus excelsa M. Bieb). Advancement in Life Sciences, 4, 20–26.
Jahan, S., Barozai, M. Y. K., Din, M., Achakzai, H., & Sajjad, A. (2017). Expressional studies of microRNAs in hepatitis B patients of Quetta, Pakistan. Pure and Applied Biology, 6, 1044–1052.
Ghani, A., Din, M., & Barozai, M. Y. K. (2018). Convergence and divergence studies of plant precursor microRNAs. Pakistan Journal of Botany, 50, 1085–1091.
Barozai, M. Y. K., & Din, M. (2017). Initial screening of plant most conserved MicroRNAs targeting infectious viruses: HBV and HCV, In 2017 14th International Bhurban Conference on Applied Sciences and Technology (IBCAST), 192–196.
Shah, S. Q., Barozai, M. Y. K., Din, M., Baloch, I. A., & Wahid, H. A. (2021). 15 RNA secondary structure analysis for abiotic stress resistant and housekeeping genes in Arabidopsis thaliana and Oryza sativa. Pure and Applied Biology, 5, 476–482.
Ambros, V., Bartel, B., Bartel, D. P., Burge, C. B., Carrington, J. C., Chen, X., Dreyfuss, G., Eddy, S. R., Griffiths-Jones, S. A. M., Marshall, M., & Matzke, M. (2003). A uniform system for microRNA annotation. RNA, 9, 277–279.
Bibi, F., Barozai, M. Y. K., & Din, M. (2017). Bioinformatics profiling and characterization of potential microRNAs and their targets in the genus Coffea. Turkish Journal of Agriculture and Forestry, 41, 191–200.
Gasparis, S., Yanushevska, Y., & Nadolska-Orczyk, A. (2017). Bioinformatic identification and expression analysis of new microRNAs from wheat (Triticum aestivum L.). Acta Physiologiae Plantarum, 39, 1–13.
Achakzai, H. K., Barozai, M. Y. K., Achakzai, A. K. K., Asghar, M., & Din, M. (2019). Profiling of 21 novel microRNA clusters and their targets in an important grain: Wheat (Triticum aestivum L.). Pakistan Journal of Botany, 51, 133–142.
Eskandarynasab, S., Roudbari, Z., & Bahreini Behzadi, M. R. (2020). Clustering based on the ontology of microRNAs target genes affecting milk production. Journal of Animal Environment, 12, 435–440.
Acknowledgements
The authors are particularly grateful to University of Balochistan, Quetta, Pakistan, and Hazara University, Mansehra, Pakistan for approving this project and facilitating the core facilities.
Funding
This research work was supported by Hazara University (HU), Mansehra, Pakistan, under number HU/699/2021.
Author information
Authors and Affiliations
Contributions
Conceptualization: WR, FM; writing-original draft preparation: WR, and IR; methodology, software, validation, formal analysis, investigation, and data curation: AB, FM, YK, AS; visualization, supervision, and project administration: AS; resources, writing-review, editing, funding acquisition, and submission: WR, DAA, ASA, SMA, FM. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
Not Applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Baqi, A., Samiullah, Rehman, W. et al. Identification and Validation of Functional miRNAs and Their Main Targets in Sorghum bicolor. Mol Biotechnol (2023). https://doi.org/10.1007/s12033-023-00988-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12033-023-00988-5