Abstract
Key message
Here, we report the decrease of JA-sensitivity and enhancement of tolerance to salt and PEG stresses in Arabidopsis overexpressing apple MdJAZ2.
Abstract
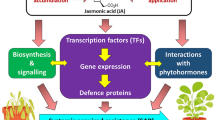
As signalling molecules, jasmonates (JAs) play significant roles in plant development and stress responses. JAZ proteins are the targets of the SCFCOI1 complex and act as the negative regulators in JA signalling pathway. However, there are no reports regarding the biological function of apple JAZ genes. In this study, one JAZ gene, MdJAZ2 from apple, was functionally characterized in detail. The expression of MdJAZ2 was up-regulated by MeJA and wounding treatments. MdJAZ2-GFP fusion protein was observed in nucleus in transient expression assay. Yeast two-hybrid and bimolecular fluorescence complementation assays revealed that MdJAZ2 could form homo- and heteromers, and also interact with F-box protein MdCOI1. Overexpression of MdJAZ2 conferred impaired JA-sensitivity in transgenic Arabidopsis, including JA-mediated root growth inhibition, susceptibility to the bacterial pathogen Pst DC3000, and the expression of JA response genes. Additionally, MdJAZ2 overexpression also improved tolerance to NaCl and PEG treatments in transgenic Arabidopsis. Together, our findings suggest that apple MdJAZ2 was not only involved in the JA response but also played roles in stress tolerance.






Similar content being viewed by others
Abbreviations
- 6-BA:
-
6-Benzylaminopurine
- AD:
-
Activation domain
- BD:
-
Binding domain
- BiFC:
-
Bimolecular fluorescence complementation
- CaMV:
-
Cauliflower mosaic virus
- COI1:
-
Coronatine insensitive 1
- CYFP:
-
C-terminal halves of the yellow fluorescent protein
- GFP:
-
Green fluorescent protein
- IAA:
-
Indole-3-acetic acid
- JA:
-
Jasmonate
- JAZ:
-
Jasmonate zim-domain
- MeJA:
-
Methyl jasmonate
- MS:
-
Murashige and skoog
- NYFP:
-
N-terminal halves of the yellow fluorescent protein
- PEG:
-
Polyethylene glycol
- Pst DC3000:
-
Pseudomonas syringae tomato DC3000
- WT:
-
Wild type
- Y2H:
-
Yeast two-hybrid
- ZIM:
-
Zinc-finger expressed in inflorescence meristem
- ZML:
-
ZIM-like
References
An XH, Tian Y, Chen KQ, Liu XJ, Liu DD, Xie XB, Cheng CG, Cong PH, Hao YJ (2015) MdMYB9 and MdMYB11 are involved in the regulation of the JA-induced biosynthesis of anthocyanin and proanthocyanidin in apples. Plant Cell Physiol 56:650–662
Bai Y, Meng Y, Huang D, Qi Y, Chen M (2011) Origin and evolutionary analysis of the plant-specific TIFY transcription factor family. Genomics 98:128–136
Browse J (2005) Jasmonate: an oxylipin signal with many roles in plants. Vitam Horm 72:431–456
Browse J, Howe GA (2008) New weapons and a rapid response against insect attack. Plant Physiol 146:832–838
Chini A, Fonseca S, Fernandez G, Adie B, Chico JM, Lorenzo O, Garcia-Casado G, Opez-Vidriero I, Lozano FM, Ponce MR, Micol JL, Solano R (2007) The JAZ family of repressors is the missing link in jasmonate signaling. Nature 448:666–671
Chini A, Fonseca S, Chico JM, Fernandez-Calvo P, Solano R (2009) The ZIM domain mediates homo- and heteromeric interactions between Arabidopsis JAZ proteins. Plant J 59:77–87
Chung HS, Howe GA (2009) A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell 21:131–145
Chung HS, Niu Y, Browse J, Howe GA (2009) Top hits in contemporary JAZ: an update on jasmonate signaling. Phytochemistry 70:1547–1559
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Devoto A, Turner JG (2005) Jasmonate-regulated Arabidopsiss tress signalling network. Physiol Plant 123:161–172
Dombrecht B, Xue GP, Sprague SJ, Kirkegaard JA, Ross JJ, Reid JB, Fitt GP, Sewelam N, Schenk PM, Manners JM, Kazan K (2007) MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19:2225–2245
Farmer EE (2007) Plant biology: jasmonate perception machines. Nature 448:659–660
Fernández-Calvo P, Chini A, Fernández-Barbero G, Chico JM, Gimenez-Ibanez S, Geerinck J, Eeckhout D, Schweizer F, Godoy M, Franco-Zorrilla JM, Pauwels L, Witters E, Puga MI, Paz-Ares J, Goossens A, Reymond P, De Jaeger G, Solano R (2011) The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23:701–715
Fonseca S, Chini A, Hamberg M, Adie B, Porzel A, Kramell R, Miersch O, Wasternack C, Solano R (2009) (+)-7-isoJasmonoyl-l-isoleucine is the endogenous bioactive jasmonate. Nat Chem Biol 5:344–350
Hakata M, Kuroda M, Ohsumi A, Hirose T, Nakamura H, Muramatsu M, Ichikawa H, Yamakawa H (2012) Overexpression of a rice TIFY gene increases grain size through enhanced accumulation of carbohydrates in the stem. Biosci Biotechnol Biochem 76:2129–2134
Hou X, Lee LYC, Xia K, Yan Y, Yu H (2010) DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev Cell 19:884–894
Howe GA, Jander G (2008) Plant immunity to insect herbivores. Annu Rev Plant Biol 59:41–66
Katagiri F, Thilmony R, He SY (2002) The Arabidopsis thaliana–Pseudomonas syringae interaction. Arabidopsis Book 2002:e0039
Katsir L, Schilmiller AL, Staswick PE, He SY, Howe GA (2008) COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc Natl Acad Sci 105:7100–7105
Li X, Yin X, Wang H, Li J, Guo C, Gao H, Zheng Y, Fan C, Wang X (2015) Genome-wide identification and analysis of the apple (Malus × domestica Borkh.) TIFY gene family. Tree Genet Genomes 11:1–13
Melotto M, Mecey C, Niu Y, Chung HS, Katsir L, Yao J, Zeng W, Thines B, Staswick P, Browse J, Howe GA, He SY (2008) A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J 55:979–988
Pauwels L, Goossens A (2011) The JAZ proteins: a crucial interface in the jasmonate signaling cascade. Plant Cell 23:3089–3100
Pauwels L, Barbero GF, Geerinck J, Tilleman S, Grunewald W, Pérez AC, Chico JM, Bossche RV, Sewell J, Gil E, García-Casado G, Witters E, Inzé D, Long JA, De Jaeger G, Solano R, Goossens A (2010) NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464:788–791
Qi T, Song S, Ren Q, Wu D, Huang H, Chen Y, Fan M, Peng W, Ren C, Xie D (2011) The jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23:1795–1814
Ren C, Pan J, Peng W, Genschik P, Hobbie L, Hellmann H, Estelle M, Gao B, Peng J, Sun C, Xie D (2005) Point mutations in Arabidopsis Cullin1 reveal its essential role in jasmonate response. Plant J 42:514–524
Seo JS, Joo J, Kim MJ, Kim YK, Nahm BH, Song SI, Cheong JJ, Lee JS, Kim JK, Choi YD (2011) OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice. Plant J 65:907–921
Shan XY, Wang JX, Chua LL, Jiang DA, Peng W, Xie DX (2011) The role of Arabidopsis rubisco activase in jasmonate-induced leaf senescence. Plant Physiol 155:751–764
Sheard LB, Tan X, Mao H, Withers J, Ben-Nissan G, Hinds TR, Kobayashi Y, Hsu F, Sharon M, Browes J, He SY, Rizo J, Howe GA, Zheng N (2010) Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468:400–405
Song S, Qi T, Huang H, Ren Q, Wu D, Chang C, Peng W, Liu Y, Peng J, Xie D (2011) The Jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect jasmonate-regulated stamen development in Arabidopsis. Plant Cell 23:1000–1013
Staswick PE (2008) JAZing up jasmonate signaling. Trends Plant Sci 13:66–71
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Thatcher LF, Cevik V, Grant M, Zhai B, Jones JDG, Manners JM, Kazan K (2016) Characterization of a JAZ7 activation-tagged Arabidopsis mutant with increased susceptibility to the fungal pathogen Fusarium oxysporum. J Exp Bot 67:2367–2386
Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J (2007) JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature 448:661–665
Vanholme B, Grunewald W, Bateman A, Kohchi T, Gheysen G (2007) The tify family previously known as ZIM. Trends Plant Sci 12:239–244
Wager A, Browse J (2012) Social network: JAZ protein interactions expand our knowledge of jasmonate signaling. Front Plant Sci 3:41
Wang Z, Cao G, Wang X, Miao J, Liu X, Chen Z, Qu LJ, Gu H (2008) Identification and characterization of COI1-dependent transcription factor genes involved in JA-mediated response to wounding in Arabidopsis plants. Plant Cell Rep 27:125–135
Wasternack C (2014) Jasmonates in plant growth and stress responses. Phytohormones: a window to metabolism, signaling and biotechnological applications. Springer, New York, pp 221–263
Wasternack C, Hause B (2013) Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann Bot 111:1021–1058
Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG (1998) COI1: an Arabidopsis gene required for jasmonate-regulated defence and fertility. Science 280:1091–1094
Xu L, Liu F, Lechner E, Genschik P, Crosby WL, Ma H, Peng W, Huang D, Xie D (2002) The SCFCOI1 ubiquitin–ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14:1919–1935
Yan Y, Stolz S, Chételat A, Reymond P, Pagni M, Dubugnon L, Farmer EE (2007) A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19:2470–2483
Yan J, Li H, Li S, Yao R, Deng H, Xie Q, Xie D (2013) The Arabidopsis F-box protein CORONATINE INSENSITIVE1 is stabilized by SCFCOI1 and degraded via the 26S proteasome pathway. Plant Cell 25:486–498
Ye H, Du H, Tang N, Li X, Xiong L (2009) Identification and expression profiling analysis of TIFY family genes involved in stress and phytohormone responses in rice. Plant Mol Biol 71:291–305
Zhang Y, Gao M, Singer SD, Fei Z, Wang H, Wang X (2012) Genome-wide identification and analysis of the TIFY gene family in grape. PLoS One 7:e44465
Zhou X, Yan S, Sun C, Li S, Li J, Xu M, Liu X, Zhang S, Zhao Q, Li Y, Fan Y, Chen R, Wang L (2015) A maize jasmonate Zim-domain protein, ZmJAZ14, associates with the JA, ABA, and GA signaling pathways in transgenic Arabidopsis. PLoS One 10(3):e0121824
Zhu Z, An F, Feng Y, Li P, Xue L, Mu A, Jiang Z, Kim JM, Kim To T, Li W, Zhang X, Yu Q, Dong Z, Chen WQ, Seki M, Zhou JM, Guo HW (2011) Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. PNAS 108:12539–12544
Zhu D, Bai X, Luo X, Chen Q, Cai H, Ji W, Zhu Y (2013) Identification of wild soybean (Glycine soja) TIFY family genes and their expression profiling analysis under bicarbonate stress. Plant Cell Rep 32:263–272
Acknowledgements
This research was supported by the Doctor Start-up Fund in Liaoning Province, China (201501031), and the Fundamental Research Funds for Central Public Welfare Research Institutes (0032015019).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
We declare that we do not have any commercial or associative interest that represents a conflict of interest in connection with the work submitted.
Additional information
Communicated by X. S. Zhang.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
An, XH., Hao, YJ., Li, EM. et al. Functional identification of apple MdJAZ2 in Arabidopsis with reduced JA-sensitivity and increased stress tolerance. Plant Cell Rep 36, 255–265 (2017). https://doi.org/10.1007/s00299-016-2077-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-016-2077-9