Abstract
Key message
Association analyses accounting for population structure and relative kinship identified eight SSR markers ( p < 0.01) showing significant association ( R 2 = 18 %) with nine agronomic traits in foxtail millet.
Abstract
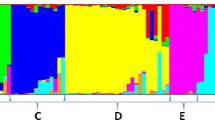
Association mapping is an efficient tool for identifying genes regulating complex traits. Although association mapping using genomic simple sequence repeat (SSR) markers has been successfully demonstrated in many agronomically important crops, very few reports are available on marker-trait association analysis in foxtail millet. In the present study, 184 foxtail millet accessions from diverse geographical locations were genotyped using 50 SSR markers representing the nine chromosomes of foxtail millet. The genetic diversity within these accessions was examined using a genetic distance-based and a general model-based clustering method. The model-based analysis using 50 SSR markers identified an underlying population structure comprising five sub-populations which corresponded well with distance-based groupings. The phenotyping of plants was carried out in the field for three consecutive years for 20 yield contributing agronomic traits. The linkage disequilibrium analysis considering population structure and relative kinship identified eight SSR markers (p < 0.01) on different chromosomes showing significant association (R 2 = 18 %) with nine agronomic traits. Four of these markers were associated with multiple traits. The integration of genetic and physical map information of eight SSR markers with their functional annotation revealed strong association of two markers encoding for phospholipid acyltransferase and ubiquitin carboxyl-terminal hydrolase located on the same chromosome (5) with flag leaf width and grain yield, respectively. Our findings on association mapping is the first report on Indian foxtail millet germplasm and this could be effectively applied in foxtail millet breeding to further uncover marker-trait associations with a large number of markers.






Similar content being viewed by others
References
Austin DF (2006) Foxtail millets (Setaria: Poaceae)—abandoned food in two hemispheres. Econ Bot 60:143–158
Barro-Kondombo C, Sagnard F, Chantereau J, Deu M, Vom BK, Durand P, Goze E, Zongo JD (2010) Genetic structure among sorghum landraces as revealed by morphological variation and microsatellites markers in three agro climatic regions of Burkina Faso. Theor Appl Genet 120:1511–1523
Barton L, Newsome SD, Chen FH, Wang H, Guilderson TP, Bettinger RL (2009) Agricultural origins and the isotopic identity of domestication in northern China. Proc Natl Acad Sci USA 106:5523–5528
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B 57:289–300
Bennetzen JL, Schmutz J, Wang H, Percifield R, Hawkins J, Pontaroli AC, Estep M, Feng L, Vaughn JN, Grimwood J et al (2012) Reference genome sequence of the model plant Setaria. Nat Biotech 30:555–561
Bohn M, Utz HF, Melchinger AE (1999) Genetic similarities among winter wheat cultivars determined on the basis of RFLPs, AFLPs, and SSRs and their use for predicting progeny variance. Crop Sci 39:228–237
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635
Breseghello F, Sorrells MS (2006) Association mapping of kernel size and milling quality in wheat (Triticum aestivum L.) cultivars. Genetics 172:1165–1177
Crossa J, Burgueno J, Dreisigacker S, Vargas M, Herrera-Foessel SA, Lillemo M, Singh RP, Trethowan R, Warburton M, Franco J et al (2007) Association analysis of historical bread wheat germplasm using additive genetic covariance of relatives and population structure. Genetics 177:1889–1913
Dekker J (2003) Evolutionary biology of the foxtail (Setaria) species-group. In: Inderjit K (ed) Principles and practices in weed management: weed biology and management. Kluwer Academic Publishers, The Netherlands, pp 65–114
Diao XM (2011) Current status of foxtail millet production in China and future development directions. The industrial production and development system of foxtail millet in china. Chinese agricultural science and technology press, Beijing, pp 20–30
Doust AN, Devos KM, Gadberry MD, Gale MD, Kellogg EA (2004) Genetic control of branching in foxtail millet. Proc Natl Acad Sci USA 101:9045–9050
Doust AN, Devos KM, Gadberry MD, Gale MD, Kellogg EA (2005) The genetic basis for inflorescence variation between foxtail and green millet (Poaceae). Genetics 169:1659–1672
Doust AN, Kellogg EA, Devos KM, Bennetzen JL (2009) Foxtail millet: a sequence-driven grass model system. Plant Physiol 149:137–141
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Falush D, Stephens M, Prithchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Guo J, Liu Y, Wang Y, Chen J, Li Y, Huang H, Qiu L, Wang Y (2012) Population structure of the wild soybean (Glycine soja) in China: implications from microsatellite analyses. Annals Bot 110:777–785
Gupta S, Kumari K, Sahu PP, Vidapu S, Prasad M (2012) Sequence-based novel genomic microsatellite markers for robust genotyping purposes in foxtail millet [Setaria italica (L.) P. Beauv.]. Plant Cell Rep 31:323–337
Hardy OJ, Vekemans X (2002) Spagedi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol 2:618–620
Hirano R, Naito K, Fukunaga K, Watanabe KN, Ohsawa R, Kawase M (2011) Genetic structure of landraces in foxtail millet (Setaria italica (L.) P. Beauv.) revealed with transposon display and interpretation to crop evolution of foxtail millet. Genome 54:498–506
Hokanson SC, Szewc-Mcfadden AK, Lamboy WF, Mcferson JR (1998) Microsatellite (SSR) markers reveal genetic identities, genetic diversity and relationships in a Malus domestica borkh. core subset collection. Theor Appl Genet 67:671–683
Hunt HV, Linden MV, Liu X, Motuzaite-Matuzeviciute G, Colledge S et al (2008) Millets across Eurasia: chronology and context of early records of the genera Panicum and Setaria from archaeological sites in the Old World. Veg History Archaeobotany 17:S5–S18
Jia X, Zhang Z, Liu Y, Zhang C, Shi Y, Song Y, Wang T, Li Y (2009) Development and genetic mapping of SSR markers in foxtail millet [Setaria italica (L.) P. Beauv.]. Theor Appl Genet 118:821–829
Jia G, Huang X, Zhi H, Zhao Y, Zhao Q, Li W, Chai Y, Yang L, Liu K, Lu H et al (2013) A haplotype map of genomic variations and genome-wide association studies of agronomic traits in foxtail millet (Setaria italica). Nat Genet 45:957–961
Jin L, Lu Y, Xiao P, Sun M, Corke H, Bao J (2010) Genetic diversity and population structure of a diverse set of rice germplasm for association mapping. Theor Appl Genet 121:475–487
Kujur A, Bajaj D, Saxena MS, Tripathi S, Upadhyaya HD, Gowda CL, Singh S, Jain M, Tyagi AK, Parida SK (2013) Functionally relevant microsatellite markers from chickpea transcription factor genes for efficient genotyping applications and trait association mapping. DNA Res 20:355–374
Kumari K, Muthamilarasan M, Misra G, Gupta S, Subramanian A, Parida SK, Chattopadhyay D, Prasad M (2013) Development of eSSR-markers in Setaria italica and their applicability in studying genetic diversity, cross-transferability and comparative mapping in millet and non-millet species. PLoS ONE 8:e67742
Lata C, Prasad M (2013) Setaria genome sequencing: an overview. J Plant Biochem Biotech 22:257–260
Lata C, Jha S, Dixit V, Sreenivasulu N, Prasad M (2011) Differential antioxidative responses to dehydration-induced oxidative stress in core set of foxtail millet cultivars [Setaria italica (L.)]. Protoplasma 248:817–828
Lata C, Gupta S, Prasad M (2013) Foxtail millet: a model crop for genetic and genomic studies in bioenergy grasses. Crit Rev Biotech 33:328–343
Li Y, Wu S (1996) Traditional maintenance and multiplication of foxtail millet (Setaria italica (L.) P. Beauv.) landraces in China. Euphytica 87:33–38
Lin HS, Chiang CY, Chang SB, Kuoh CS (2011) Development of simple sequence repeats (SSR) markers in Setaria italica (Poaceae) and cross-amplification in related species. Int J Mol Sci 12:7835–7845
Liu K, Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21:2128–2129
Liu K, Goodman M, Muse S, Smith JS, Buckler E, Doebley J (2003) Genetic structure and diversity among maize inbred lines as inferred from DNA microsatellites. Genetics 165:2117–2128
Liu L, Wang L, Yao J, Zheng J, Zhao C (2010) Association mapping of six agronomic traits on chromosome 4A of wheat (Triticum aestivum L.). Mol Plant. doi:10.5376/mpb.2010.01.0005
Liu Z, Bai G, Zhang D, Zhu C, Xia X, Cheng R, Shi Z (2011) Genetic diversity and population structure of elite foxtail millet [Setaria italica (L.) P. Beauv.] germplasm in China. Crop Sci 51:1655–1663
Lu H, Zhang J, Liu KB, Wu N, Li Y et al (2009) Earliest domestication of common millet (Panicum miliaceum) in east Asia extended to 10,000 years ago. Proc Natl Acad Sci USA 106:7367–7372
Maccaferri M, Sanguineti MC, Noli E, Tuberosa R (2005) Population structure and long-range linkage disequilibrium in a durum wheat elite collection. Mol Breed 15:271–289
Maccaferri M, Sanguineti MC, Demontis A et al (2011) Association mapping in durum wheat grown across a broad range of water regimes. J Exp Bot 62:409–438
Mather DE, Hayes PM, Chalmers KJ, Eglinton JK, Matus I, Richardson KL, Von-Zitzewitz J, Marquez- Cedillo L, Hearnden P, Pal N (2004) Use of SSR marker data to study linkage disequilibrium and population structure in Hordeum vulgare: prospects for association mapping in barley. In: Spunar J, Janikova J (eds) Proceedings from the 9th international barley genetics symposium. Czech J Genet Plant Breed, Czech Republic, pp 302–307
Mauro-Herrera M, Wang X, Barbier H, Brutnell TP, Devos KM, Doust AN (2013) Genetic control and comparative genomic analysis of flowering time in Setaria (Poaceae). G3 3:283–295
Moon YK, Hong JP, Cho YC, Yang SJ, An G, Kim WT (2009) Structure and expression of OsUBP6, an ubiquitin-specific protease 6 homolog in rice (Oryza sativa L.). Mol Cells 28:463–472
Muthamilarasan M, Suresh BV, Pandey G, Kumari K, Parida SK, Prasad M (2013) Development of 5123 Intron length polymorphic (ILP) markers for large-scale genotyping applications in foxtail millet [Setaria italica (L.)]. DNA Res. doi:10.1093/dnares/dst039
Pandey G, Misra G, Kumari K, Gupta S, Parida SK, Chattopadhyay D, Prasad M (2013) Genome-wide development and use of microsatellite markers for large-scale genotyping applications in foxtail millet [Setaria italica (L.)]. DNA Res 20:197–207
Pejic I, Ajmone-Marsan P, Morgante M, Kozumplick V, Castiglioni P, Taramino G, Motto M (1998) Comparative analysis of genetic similarity among maize inbred lines detected by RFLPs, RAPDs, SSRs, and AFLPs. Theor Appl Genet 97:1248–1255
Prasad M, Varshney RK, Roy JK, Balyan HS, Gupta PK (2000) The use of microsatellites for detecting DNA polymorphism, genotype identification and genetic diversity in wheat. Theor Appl Genet 100:584–592
Pritchard JK, Przeworski M (2001) Linkage disequilibrium in humans models data. Am J Human Genet 69:1–14
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Razafinarivo NJ, Guyot R, Davis AP, Couturon E, Hamon S, Crouzillat D, Rigoreau M, Dubreuil-Tranchant C, Poncet V, De-Kochko A et al (2013) Genetic structure and diversity of coffee (Coffea) across Africa and the Indian Ocean islands revealed using microsatellites. Annals Bot 111:229–248
Reimer SO, Pozniak CJ, Clarke FR, Clarke JM, Somers DJ, Knox RE, Singh AK (2008) Association mapping of yellow pigment in an elite collection of durum wheat cultivars and breeding lines. Genome 51:1016–1025
Rice WR (1989) Analyzing tables of statistical tests. Evolution 43:223–225
Rogers JS (1972) Measures of genetic similarity and genetic distance. Studies in genetics VII. University Texas Publication, Texas, pp 145–153
Saghai-Maroof MA, Biyaschev RM, Yang GP, Zhang Q, Allard RW (1994) Extaordinary polymorphism microsatellite DNA in barley: species diversity, chromosomal location and population dynamics. Proc Natl Acad Sci USA 91:5466–5470
Sneath PHA, Sokal RR (1973) Numerical taxonomy: the principles and practice of and numerical classification. Freeman, San Francisco, p 573
Suresh BV, Muthamilarasan M, Misra G, Prasad M (2013) FmMDb: a versatile database of foxtail millet markers for millets and bioenergy grasses research. PLoS ONE 8:e71418
Tsuruta SI, Hashiguchi M, Ebina M, Matsuo T, Yamamoto T, Kobayashi M, Takahara M, Nakagawa H, Akashi R (2005) Development and characterization of simple sequence repeat markers in Zoysia japonica Steud. Grassland Sci 51:249–257
Upadhyaya HD, Wang YH, Sharma S, Singh S, Hasenstein KH (2012) SSR markers linked to kernel weight and tiller number in sorghum identified by association mapping. Euphytica 187:401–410
van Oosterhout C, Hutchinson W, Wills D, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol 4:535–538
Vavilov NI (1926) Studies on the origin of cultivated plants. Bull Appl Bot Plant Breed 16:139–248
Wang C, Jia G, Zhi H, Niu Z, Chai Y, Li W, Wang Y, Li H, Lu P, Zhao B, Diao X (2012) Genetic diversity and population structure of chinese foxtail millet [Setaria italica (L.) Beauv.] landraces. G3 Genes Genomics Genetics 2:769–777
Weir BS, Hill WG (2002) Estimating F-statistics. Ann Rev Genetics 36:721–750
Wong YC, Ho CL, Kulaveerasingam H, Zain AM, Napis S, Zaman FQ (2007) Analyses of expressed sequence tags (ESTs) from panicles of the indica rice cultivar MR84 during grain filling stages and molecular characterisation of ADP-glucose pyrophosphorylase small subunit. Asia Pacific J Mol Biol Biotech 15:81–90
Yan WG, Li Y, Agrama HA, Luo D, Gao F, Lu X, Ren G (2009) Association mapping of stigma and spikelet characteristics in rice (Oryza sativa L.). Mol Breed 24:277–292
Yu J, Buckler ES (2006) Genetic association mapping and genome organization of maize. Curr Opin Biotech 17:1–6
Zhang D, Zhang H, Wang M, Sun J, Qi Y, Wang F, Wei X, Han L, Wang X, Li Z (2009a) Genetic structure and differentiation of Oryza sativa L. in China revealed by microsatellites. Theor Appl Genet 119:1105–1117
Zhang M, Fan J, Taylor DC, Ohlrogge JB (2009b) DGAT1 and PDAT1 acyltransferases have overlapping functions in Arabidopsis triacylglycerol biosynthesis and are essential for normal pollen and seed development. Plant Cell 21:3885–3901
Zhang G, Liu X, Quan Z, Cheng S, Xu X, Pan S, Xie M, Zeng P, Yue Z, Wang W (2012) Genome sequence of foxtail millet (Setaria italica) provides insights into grass evolution and biofuel potential. Nat Biotech 30:549–554
Zondervan KT, Cardon LR (2004) The complex interplay among factors that influence allelic association. Nat Rev Genetics 5:89–100
Zoric M, Dejan D, Kobiljski B, Quarrie S, Barnes J (2012) Population structure in a wheat core collection and genomic loci associated with yield under contrasting environments. Genetica 140:259–275
Acknowledgments
Thanks are due to the Director, National Institute of Plant Genome Research (NIPGR), New Delhi, India for providing facilities. This work area was supported by the core grant of NIPGR. Dr. Sarika Gupta and Mr. Mehanathan Muthamilarasan acknowledge the award of DST-Young Scientist and Junior Research Fellowship from the Dept. of Science and Technology (DST) and University Grants Commission, New Delhi, respectively. We are also thankful to the National Bureau of Plant Genetic Resources, New Delhi/Hyderabad/Akola, India for providing the seed materials.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Dhingra.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gupta, S., Kumari, K., Muthamilarasan, M. et al. Population structure and association mapping of yield contributing agronomic traits in foxtail millet. Plant Cell Rep 33, 881–893 (2014). https://doi.org/10.1007/s00299-014-1564-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-014-1564-0