Abstract
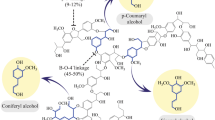
Many bacterial strains have been demonstrated to biodegrade lignin for contaminant removal or resource regeneration. The goal of this study was to investigate the biodegradation amount and associated pathways of three lignin monomers, vanillic, p-coumaric, and syringic acid by strain Sphingobacterium sp. HY-H. Vanillic, p-coumaric, and syringic acid degradation with strain HY-H was estimated as 88.71, 76.67, and 72.78%, respectively, after 96 h. Correspondingly, the same three monomers were associated with a COD removal efficiency of 87.30, 55.17, and 67.23%, and a TOC removal efficiency of 82.14, 61.03, and 43.86%. The results of GC–MS, HPLC, FTIR, and enzyme activities show that guaiacol and o-dihydroxybenzene are key intermediate metabolites of the vanillic acid and syringic acid degradation. p-Hydroxybenzoic acid is an important intermediate metabolite for p-coumaric and syringic acid degradation. LiP and MnP play an important role in the degradation of lignin monomers and their intermediate metabolites. One possible pathway is that strain HY-H degrades lignin monomers into guaiacol (through decarboxylic and demethoxy reaction) or p-hydroxybenzoic acid (through side-chain oxidation); then guaiacol demethylates to o-dihydroxybenzene. The p-hydroxybenzoic acid and o-dihydroxybenzene are futher through ring cleavage reaction to form small molecule acids (butyric, valproic, oxalic acid, and propionic acid) and alcohols (ethanol and ethanediol), then these acids and alcohols are finally decomposed into CO2 and H2O through the tricarboxylic acid cycle. If properly optimized and controlled, the strain HY-H may play a role in breaking down lignin-related compounds for biofuel and chemical production.





Similar content being viewed by others
References
Ayyachamy M, Cliffe FE, Coyne JM et al (2013) Lignin: untapped biopolymers in biomass conversion technologies. Biomass Convers Biorefin 3(3):255–269
Meng X, Ragauskas AJ (2014) Recent advances in understanding the role of cellulose accessibility in enzymatic hydrolysis of lignocellulosic substrates. Curr Opin Biotechnol 27(6):150–158
Camarero S, Martínez MJ, Martínez AT (2014) Understanding lignin biodegradation for the improved utilization of plant biomass in modern biorefineries. Biofuels Bioprod Biorefin 8(5):615–625
Beckham GT, Johnson CW, Karp EM et al (2016) Opportunities and challenges in biological lignin valorization. Curr Opin Biotechnol 42:40–53
Brzonova I, Kozliak E, Kubátová A et al (2014) Kenaf biomass biodecomposition by basidiomycetes and actinobacteria in submerged fermentation for production of carbohydrates and phenolic compounds. Bioresour Technol 173:352–360
Baltierra-Trejo E, Sánchez-Yáñez JM et al (2015) Production of short-chain fatty acids from the biodegradation of wheat straw lignin by Aspergillus fumigatus. Bioresour Technol 196:418–425
Xie SX, Qin X, Cheng YB et al (2015) Simultaneous conversion of all cell wall components by an oleaginous fungus without chemi-physical pretreatment. Green Chem 17:1657–1667
Zou L, Ross BM, Hutchison LJ et al (2015) Fungal demethylation of Kraft lignin. Enzyme Microb Technol 74:44–50
Sainsbury PD, Hardiman EM, Ahmad M et al (2013) Breaking down lignin to high-value chemicals: the conversion of lignocellulose to vanillin in a gene deletion mutant of Rhodococcus jostii RHA1. Chem Biol 8:2151–2156
Mycroft Z, Gomis M, Mines P et al (2015) Biocatalytic conversion of lignin to aromatic dicarboxylic acids in Rhodococcus jostii RHA1 by re-routing aromatic degradation pathways. Green Chem 17:4975–4979
Sainsbury PD, Mineyeva Y, Mycroft Z et al (2015) Chemical intervention in bacterial lignin degradation pathways: development of selective inhibitors for intradiol and extradiol catechol dioxygenases. Bioorg Chem 60(4):102–109
Mathews SL, Grunden AM, Pawlak J (2016) Degradation of lignocellulose and lignin by Paenibacillus glucanolyticus. Int Biodeterior Biodegrad 110:79–86
Suman SK, Dhawaria M, Tripathi D et al (2016) Investigation of lignin biodegradation by Trabulsiella sp. isolated from termite gut. Int Biodeterior Biodegrad 112:12–17
Huang X-F, Santhanam N, Badri DV et al (2013) Isolation and characterization of lignin-degrading bacteria from rainforest soils. Biotechnol Bioeng 110(6):1616–1626
Rashid GMM, Taylor CR, Liu Y et al (2015) Identification of manganese superoxide dismutase from Sphingobacterium sp. T2 as a novel bacterial enzyme for lignin oxidation. Chem Biol 10:2286–2294
Taylor CR, Hardiman EM, Ahmad M et al (2012) Isolation of bacterial strains able to metabolize lignin from screening of environmental samples. J Appl Microbiol 113:521–530
Rashid GM, Taylor CR, Liu Y, Zhang X, Rea D et al (2015) Identification of manganese superoxide dismutase from Sphingobacterium sp. T2 as a novel bacterial enzyme for lignin oxidation. Chem Biol 10:2286–2294
Li A, Ngo TPN, Yan J et al (2012) Whole-cell based solvent-free system for one-pot production of biodiesel from waste grease. Bioresour Technol 114:725–729
Dongqi W, Yishan L, Wenjing D et al (2013) Optimization and characterization of lignosulfonate biodegradation process by a bacterial strain, Sphingobacterium sp. HY-H. Int Biodeterior Biodegrad 85:365–371
Nakagawa Y, Sakamoto Y, Kikuchi S et al (2010) A chimeric laccase with hybrid properties of the parental Lentinula edodes laccase. Microbiol Res 165:392–401
Orth AB, Royse DJ, Tien M (1993) Ubiquity of lignin-degrading peroxidases among various wood-degrading fungi. Appl Environ Microbiol 59(12):4017–4023
Kapich AN, Prior BA, Botha A et al (2004) Effect of lignocellulose-containing substrates on production of ligninolytic peroxidases in submerged cultures of Phanerochaete chrysosporium ME-446. Enzyme Microb Technol 34:187–195
Sun S, Chen Y, Li L (2016) Enhanced degradation of lignin by the strain fusion of Pseudomonas putida and Gordonia sp. Biocatalysis 34(4):144–151
Shi Y, Chai L, Tang C et al (2013) Biochemical investigation of kraft lignin degradation by Pandoraea sp. B-6 isolated from bamboo slips. Bioprocess Biosyst Eng 36(12):1957–1965
Shi Y, Chai L, Tang C et al (2013) Characterization and genomic analysis of kraft lignin biodegradation by the beta-proteobacterium Cupriavidus basilensis B-8. Biotechnol Biofuels 6:1–14
Kumar M, Singh J, Kumar M, Singh et al (2015) Investigating the degradation process of kraft lignin by β-proteobacterium, Pandoraea sp. ISTKB. Environ Sci Pollut Res 22:15690–15702
Ravi K, García-Hidalgo J, Gorwa-Grauslund MF et al (2017) Conversion of lignin model compounds by Pseudomonas putida KT2440 and isolates from compost. Appl Microbiol Biotechnol 101:5059–5070
Yadav S, Chandra R (2015) Syntrophic co-culture of Bacillus subtilis and Klebsiella pneumonia for degradation of kraft lignin discharged from rayon grade pulp industry. J Environ Sci 33:229–238
Colbert CL, Agar NYR, Kumar P et al (2013) Structural characterization of Pandoraea pnomenusa B-356 biphenyl dioxygenase reveals features of potent polychlorinated biphenyl-degrading enzymes: crystal structure of a potent PCB. Degrad Enzyme 8(1):52–55
Raj A, Krishna Reddy MM, Chandra R (2007) Decolourisation and treatment of pulp and paper mill effluent by lignin-degrading Bacillus sp. J Chem Technol Biotechnol 82:399–406
Masai E, Katayama Y, Fukuda M (2007) Genetic and biochemical investigations on bacterial catabolic pathways for lignin-derived aromatic compounds. Biosci Biotechnol Biochem 71(1):1–15
Rahouti M, Steiman R, Seigle-Murandi F et al (1999) Growth of 1044 strains and species of fungi on 7 phenolic lignin model compounds. Chemosphere 38:2549–2559
Yang XE, Ma FY, Zeng YL et al (2010) Structure alteration of lignin in corn stover degraded by white-rot fungus Irpex lacteus CD2. Int Biodeterior Biodegrad 64(2):119–123
Priyadarshinee R, Kumar A, Mandal T, Dasguptamandal D (2017) Exploring the diverse potentials of Planococcus sp. TRC1 for the deconstruction of recalcitrant kraft lignin. Int J Environ Sci 14(8):1–16
He Y, Li X, Ben H et al (2017) Lipid production from dilute alkali corn stover lignin by Rhodococcus strains. Sustain Chem Eng 5:2302–2311
Hettiaratchi JPA, Jayasinghe PA, Bartholameuz EM et al (2014) Waste degradation and gas production with enzymatic enhancement in anaerobic and aerobic landfill bioreactors. Bioresour Technol 159:433–436
Wen X, Jia Y, Li J (2009) Degradation of tetracycline and oxytetracycline by crude lignin peroxidase prepared from Phanerochaete chrysosporium–a white rot fungus. Chemosphere 75:1003–1007
Zheng Y, Chai L, Yang Z et al (2013) Characterization of a newly isolated bacterium Pandoraea sp. B-6 capable of degrading kraft Lignin. J Cent South Univ 20(3):757–763
Cserháti M, Kriszt B, Szoboszlay S et al (2012) De dovo genome project of Cupriavidus basilensis OR16. J Bacteriol 194(8):2109–2110
Zhang ZY, Pan LP, Li HH (2010) Isolation, identification and characterization of soil microbes which degrade phenolic allelochemicals. J Appl Microbiol 108:1839–1849
Chen SY, Guo LY, Bai JG et al (2015) Biodegradation of p-hydroxybenzoic acid in soil by Pseudomonas putida CSY-P1 isolated from cucumber rhizosphere soil. Plant Soil 389(1–2):197–210
Inderjit ETN (2003) Bioassays and field studies for allelopathy in terrestrial plants: progress and problems. Crit Rev Plant Sci 22:221–238
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Wang, J., Liang, J. & Gao, S. Biodegradation of Lignin Monomers Vanillic, p-Coumaric, and Syringic Acid by the Bacterial Strain, Sphingobacterium sp. HY-H. Curr Microbiol 75, 1156–1164 (2018). https://doi.org/10.1007/s00284-018-1504-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-018-1504-2