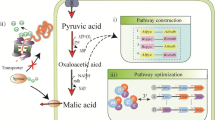
Abstract
Efficient transporters are necessary for high concentration and purity of desired products during industrial production. In this study, we explored the mechanism of substrate transport and preference of the C4-dicarboxylic acid transporter AoMAE in the fungus Myceliophthora thermophila, and investigated the roles of 18 critical amino acid residues within this process. Among them, the residue Arg78, forming a hydrogen bond network with Arg23, Phe25, Thr74, Leu81, His82, and Glu94 to stabilize the protein conformation, is irreplaceable for the export function of AoMAE. Furthermore, varying the residue at position 100 resulted in changes to the size and shape of the substrate binding pocket, leading to alterations in transport efficiencies of both malic acid and succinic acid. We found that the mutation T100S increased malate production by 68%. Using these insights, we successfully generated an AoMAE variant with mutation T100S and deubiquitination, exhibiting an 81% increase in the selective export activity of malic acid. Simply introducing this version of AoMAE into M. thermophila wild-type strain increased production of malic acid from 1.22 to 54.88 g/L. These findings increase our understanding of the structure–function relationships of organic acid transporters and may accelerate the process of engineering dicarboxylic acid transporters with high efficiency.
Key points
• This is the first systematical analysis of key residues of a malate transporter in fungi.
• Protein engineering of AoMAE led to 81% increase of malate export activity.
• Arg78 was essential for the normal function of AoMAE in M. thermophila.
• Substitution of Thr100 affected export efficiency and substrate selectivity of AoMAE.










Similar content being viewed by others
Data availability
All data generated during this study are included in this published article (and its supplementary material file).
References
Ahn JH, Jang Y-S, Lee SY (2016) Production of succinic acid by metabolically engineered microorganisms. Curr Opin Biotechnol 42:54–66. https://doi.org/10.1016/j.copbio.2016.02.034
Alonso S, Rendueles M, Díaz M (2015) Microbial production of specialty organic acids from renewable and waste materials. Crit Rev Biotechnol 35(4):497–513. https://doi.org/10.3109/07388551.2014.904269
Berka RM, Grigoriev IV, Otillar R, Salamov A, Grimwood J, Reid I, Ishmael N, John T, Darmond C, Moisan MC, Henrissat B, Coutinho PM, Lombard V, Natvig DO, Lindquist E, Schmutz J, Lucas S, Harris P, Powlowski J, Bellemare A, Taylor D, Butler G, de Vries RP, Allijn IE, van den Brink J, Ushinsky S, Storms R, Powell AJ, Paulsen IT, Elbourne LD, Baker SE, Magnuson J, Laboissiere S, Clutterbuck AJ, Martinez D, Wogulis M, de Leon AL, Rey MW, Tsang A (2011) Comparative genomic analysis of the thermophilic biomass-degrading fungi Myceliophthora thermophila and Thielavia terrestris. Nat Biotechnol 29(10):922–927. https://doi.org/10.1038/nbt.1976
Brown SH, Bashkirova L, Berka R, Chandler T, Doty T, McCall K, McCulloch M, McFarland S, Thompson S, Yaver D, Berry A (2013) Metabolic engineering of Aspergillus oryzae NRRL 3488 for increased production of L-malic acid. Appl Microbiol Biotechnol 97(20):8903–8912. https://doi.org/10.1007/s00253-013-5132-2
Caballero D, Virrueta A, O’Hern CS, Regan L (2016) Steric interactions determine side-chain conformations in protein cores. Protein Eng Des Sel 29(9):367–376. https://doi.org/10.1093/protein/gzw027
Cao W, Yan L, Li M, Liu X, Xu Y, Xie Z, Liu H (2020) Identification and engineering a C4-dicarboxylate transporter for improvement of malic acid production in Aspergillus niger. Appl Microbiol Biotechnol 104(22):9773–9783. https://doi.org/10.1007/s00253-020-10932-1
Chen YH, Hu L, Punta M, Bruni R, Hillerich B, Kloss B, Rost B, Love J, Siegelbaum SA, Hendrickson WA (2010) Homologue structure of the SLAC1 anion channel for closing stomata in leaves. Nature 467(7319):1074–1080. https://doi.org/10.1038/nature09487
Chen X, Xu G, Xu N, Zou W, Zhu P, Liu L, Chen J (2013) Metabolic engineering of Torulopsis glabrata for malate production. Metab Eng 19:10–16. https://doi.org/10.1016/j.ymben.2013.05.002
Chen J, Zhu X, Tan Z, Xu H, Tang J, Xiao D, Zhang X (2014) Activating C4-dicarboxylate transporters DcuB and DcuC for improving succinate production. Appl Microbiol Biotechnol 98(5):2197–2205. https://doi.org/10.1007/s00253-013-5387-7
Chen X, Wang Y, Dong X, Hu G, Liu L (2017) Engineering rTCA pathway and C4-dicarboxylate transporter for L-malic acid production. Appl Microbiol Biotechnol 101(10):4041–4052. https://doi.org/10.1007/s00253-017-8141-8
Chenna R, Sugawara H, Fau - Koike T, Koike T Fau - Lopez R, Lopez R Fau - Gibson TJ, Gibson Tj Fau - Higgins DG, Higgins Dg Fau - Thompson JD, Thompson JD (2003) Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31(13):3497–3500
Combs SA, Deluca SL, Deluca SH, Lemmon GH, Nannemann DP, Nguyen ED, Willis JR, Sheehan JH, Meiler J (2013) Small-molecule ligand docking into comparative models with Rosetta. Nat Protoc 8(7):1277–1198. https://doi.org/10.1038/nprot.2013.074
Dai Z, Zhou H, Zhang S, Gu H, Yang Q, Zhang W, Dong W, Ma J, Fang Y, Jiang M, Xin F (2018) Current advance in biological production of malic acid using wild type and metabolic engineered strains. Bioresour Technol 258:345–353. https://doi.org/10.1016/j.biortech.2018.03.001
Darbani B, Stovicek V, van der Hoek SA, Borodina I (2019) Engineering energetically efficient transport of dicarboxylic acids in yeast Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 116(39):19415–19420. https://doi.org/10.1073/pnas.1900287116
Diallinas G (2021) Transporter specificity: a tale of loosened elevator-sliding. Trends Biochem Sci 46(9):708–717. https://doi.org/10.1016/j.tibs.2021.03.007
Foot N, Henshall T, Kumar S (2017) Ubiquitination and the regulation of membrane proteins. Physiol Rev 97(1):253–281. https://doi.org/10.1152/physrev.00012.2016
Goldberg I, Rokem JS, Pines O (2006) Organic acids: old metabolites, new themes. J Chem Technol Biotechnol 81(10):1601–1611. https://doi.org/10.1002/jctb.1590
Grobler J, Bauer F, Subden RE, Vuuren HJJV (1995) The mae1 gene of Schizosaccharomyces pombe encodes a permease for malate and other C4 dicarboxylic acids. Yeast 11(15):1485–1491. https://doi.org/10.1002/YEA.320111503
Groeneveld M, Weme RG, Duurkens RH, Slotboom DJ (2010) Biochemical characterization of the C4-dicarboxylate transporter DctA from Bacillus subtilis. J Bacteriol 192(11):2900–2907. https://doi.org/10.1128/JB.00136-10
Jones DT, Fau TW, Thornton -, JM, Thornton JM, (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8(3):275–282
Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, Bridgland A, Meyer C, Kohl SAA, Ballard AJ, Cowie A, Romera-Paredes B, Nikolov S, Jain R, Adler J, Back T, Petersen S, Reiman D, Clancy E, Zielinski M, Steinegger M, Pacholska M, Berghammer T, Bodenstein S, Silver D, Vinyals O, Senior AW, Kavukcuoglu K, Kohli P, Hassabis D (2021) Highly accurate protein structure prediction with AlphaFold. Nature 596:583–589. https://doi.org/10.1038/s41586-021-03819-2
Kosti V, Papageorgiou I, Diallinas G (2010) Dynamic elements at both cytoplasmically and extracellularly facing sides of the UapA transporter selectively control the accessibility of substrates to their translocation pathway. J Mol Biol 397(5):1132–1143. https://doi.org/10.1016/j.jmb.2010.02.037
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–4. https://doi.org/10.1093/molbev/msw054
Li J, Lin L, Sun T, Xu J, Ji J, Liu Q, Tian C (2020a) Direct production of commodity chemicals from lignocellulose using Myceliophthora thermophila. Metab Eng 61:416–426. https://doi.org/10.1016/j.ymben.2019.05.007
Li J, Zhang Y, Li J, Sun T, Tian C (2020b) Metabolic engineering of the cellulolytic thermophilic fungus Myceliophthora thermophila to produce ethanol from cellobiose. Biotechnol Biofuels 13:23. https://doi.org/10.1186/s13068-020-1661-y
Liu J, Xie Z, Shin H-D, Li J, Du G, Chen J, Liu L (2017a) Rewiring the reductive tricarboxylic acid pathway and L-malate transport pathway of Aspergillus oryzae for overproduction of L-malate. J Biotechnol 253:1–9. https://doi.org/10.1016/j.jbiotec.2017.05.011
Liu Q, Gao R, Li J, Lin L, Zhao J, Sun W, Tian C (2017b) Development of a genome-editing CRISPR/Cas9 system in thermophilic fungal Myceliophthora species and its application to hyper-cellulase production strain engineering. Biotechnol Biofuels 10:1. https://doi.org/10.1186/s13068-016-0693-9
Liu J, Li J, Shin HD, Du G, Chen J, Liu L (2018) Biological production of L-malate: recent advances and future prospects. World J Microbiol Biotechnol 34(1):6. https://doi.org/10.1007/s11274-017-2349-8
Ma C, Remani S, Kotaria R, Mayor JA, Walters DE, Kaplan RS (2006) The mitochondrial citrate transport protein: evidence for a steric interaction between glutamine 182 and leucine 120 and its relationship to the substrate translocation pathway and identification of other mechanistically essential residues. Biochim Biophys Acta 1757(9–10):1271–1276. https://doi.org/10.1016/j.bbabio.2006.06.011
MacGurn JA, Hsu PC, Emr SD (2012) Ubiquitin and membrane protein turnover: from cradle to grave. Annu Rev Biochem 81:231–259. https://doi.org/10.1146/annurev-biochem-060210-093619
O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR (2011) Open Babel: An open chemical toolbox. J Cheminformatics 3(1):33. https://doi.org/10.1186/1758-2946-3-33
Osawa H, Matsumoto H (2006) Cytotoxic thio-malate is transported by both an aluminum-responsive malate efflux pathway in wheat and the MAE1 malate permease in Schizosaccharomyces pombe. Planta 224:462–471
Park H, Bradley P, Greisen P Jr, Liu Y, Mulligan VK, Kim DE, Baker D, DiMaio F (2016) Simultaneous optimization of biomolecular energy functions on features from small molecules and macromolecules. J Chem Theory Comput 12(12):6201–6212. https://doi.org/10.1021/acs.jctc.6b00819
Quick M, Abramyan AM, Wiriyasermkul P, Weinstein H, Shi L, Javitch JA (2018) The LeuT-fold neurotransmitter: sodium symporter MhsT has two substrate sites. Proc Natl Acad Sci U S A 115(34):E7924–E7931. https://doi.org/10.1073/pnas.1717444115
Sauer M, Porro D, Mattanovich D, Branduardi P (2008) Microbial production of organic acids: expanding the markets. Trends Biotechnol 26(2):100–108. https://doi.org/10.1016/j.tibtech.2007.11.006
Sebastian J, Hegde K, Kumar P, Rouissi T, Brar SK (2019) Bioproduction of fumaric acid: an insight into microbial strain improvement strategies. Crit Rev Biotechnol 39(6):817–834. https://doi.org/10.1080/07388551.2019.1620677
Soares-Silva I, Sa-Pessoa J, Myrianthopoulos V, Mikros E, Casal M, Diallinas G (2011) A substrate translocation trajectory in a cytoplasm-facing topological model of the monocarboxylate/H+ symporter Jen1p. Mol Microbiol 81(3):805–817. https://doi.org/10.1111/j.1365-2958.2011.07729.x
Soares-Silva I, Ribas D, Foskolou IP, Barata B, Bessa D, Paiva S, Queiros O, Casal M (2015) The Debaryomyces hansenii carboxylate transporters Jen1 homologues are functional in Saccharomyces cerevisiae. FEMS Yeast Res 15(8): fov094. https://doi.org/10.1093/femsyr/fov094
Thakker C, Martínez I, Li W, San K-Y, Bennett GN (2015) Metabolic engineering of carbon and redox flow in the production of small organic acids. J Ind Microbiol Biotechnol 42(3):403–422. https://doi.org/10.1007/s10295-014-1560-y
Valentini M, Storelli N, Lapouge K (2011) Identification of C4-dicarboxylate transport systems in Pseudomonas aeruginosa PAO1. J Bacteriol 193(17):4307–4316. https://doi.org/10.1128/JB.05074-11
Vlanti A, Amillis S, Koukaki M, Diallinas G (2006) A novel-type substrate-selectivity filter and ER-exit determinants in the UapA purine transporter. J Mol Biol 357(3):808–819
Wang H, Sun T, Zhao Z, Gu S, Liu Q, Wu T, Wang D, Tian C, Li J (2021) Transcriptional profiling of Myceliophthora thermophila on galactose and metabolic engineering for improved galactose utilization. Front Microbiol 12:664011. https://doi.org/10.3389/fmicb.2021.664011
Wei L, Liu J, Qi H, Wen J (2015) Engineering Scheffersomyces stipitis for fumaric acid production from xylose. Bioresour Technol 187:246–254. https://doi.org/10.1016/j.biortech.2015.03.122
Wu N, Zhang J, Chen Y, Xu Q, Song P, Li Y, Li K, Liu H (2022) Recent advances in microbial production of L-malic acid. Appl Microbiol Biotechnol. https://doi.org/10.1007/s00253-022-12260-y
Xie S, Shen B, Zhang C, Huang X, Zhang Y (2014) sgRNAcas9: a software package for designing CRISPR sgRNA and evaluating potential off-target cleavage sites. PLoS ONE 9(6):e100448. https://doi.org/10.1371/journal.pone.0100448
Xu J, Li J, Lin L, Liu Q, Sun W, Huang B, Tian C (2015) Development of genetic tools for Myceliophthora thermophila. BMC Biotechnol 15:35. https://doi.org/10.1186/s12896-015-0165-5
Yang L, Christakou E, Vang J, Lübeck M, Lübeck PS (2017) Overexpression of a C4-dicarboxylate transporter is the key for rerouting citric acid to C4-dicarboxylic acid production in Aspergillus carbonarius. Microb Cell Fact 16(1):43. https://doi.org/10.1186/s12934-017-0660-6
Yang YJ, Liu Y, Liu DD, Guo WZ, Wang LX, Wang XJ, Lv HX, Yang Y, Liu Q, Tian CG (2022) Development of a flow cytometry-based plating-free system for strain engineering in industrial fungi. Appl Microbiol Biotechnol 106(2):713–727. https://doi.org/10.1007/s00253-021-11733-w
Zelle RM, de Hulster E, van Winden WA, de Waard P, Dijkema C, Winkler AA, Geertman JM, van Dijken JP, Pronk JT, van Maris AJ (2008) Malic acid production by Saccharomyces cerevisiae: engineering of pyruvate carboxylation, oxaloacetate reduction, and malate export. Appl Environ Microbiol 74(9):2766–2777. https://doi.org/10.1128/AEM.02591-07
Zhang W, Bell EW, Yin M, Zhang Y (2020) EDock: blind protein-ligand docking by replica-exchange monte carlo simulation. J Cheminform 12(1):37. https://doi.org/10.1186/s13321-020-00440-9
Zientz E, Janausch IG, Six S, Unden G (1999) Functioning of DcuC as the C4-dicarboxylate carrier during glucose fermentation by Escherichia coli. J Bacteriol 181(12):3716–3720
Funding
This work was supported by funding from the Key Project of the Ministry of Science and Technology of China (2018YFA0901400 and 2018YFA0900500), the National Natural Science Foundation of China (32071424, 31972878), the Chinese Academy of Sciences (XDA21060900), the Tianjin Synthetic Biotechnology Innovation Capacity Improvement Project (TSBICIP-CXRC-022 and TSBICIP-KJGG-015) and the Youth Innovation Promotion Association of the Chinese Academy of Sciences (2020183).
Author information
Authors and Affiliations
Contributions
LJ and WT conceived and designed research. WT conducted experiments and analyzed data. YW contributed software and analyzed data. WT wrote the manuscript. TC and LJ reviewed and edited the manuscript. All authors read and approved the manuscript.
Corresponding authors
Ethics declarations
Ethics approval
This article does not contain any studies with human participants performed by any of the authors.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wu, T., Wang, Y., Li, J. et al. Dissecting key residues of a C4-dicarboxylic acid transporter to accelerate malate export in Myceliophthora. Appl Microbiol Biotechnol 107, 609–622 (2023). https://doi.org/10.1007/s00253-022-12336-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-022-12336-9