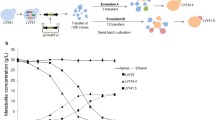
Abstract
Production of ethanol from xylose by recombinant Saccharomyces cerevisiae is suboptimal with slow fermentation rate, compared with that from glucose. In this study, a strain-expressing Scheffersomyces stipitis xylose reductase–xylitol dehydrogenase (XR-XDH) pathway was subjected to adaptive evolution on xylose; this approach generated populations with the significantly improved cell growth and ethanol production rate. Mutants were isolated, and the best one was used for sporulation to generate eight stable mutant strains with improved xylose fermentation ability. They were used in a microarray assay to study the molecular basis of the enhanced phenotype. The enriched transcriptional differences among the eight mutant strains and the native strain revealed novel responses to xylose, which likely contributes to the improved xylose utilization. The upregulated vitamin B1 and B6 biosynthesis indicated that thiamine served as an important cofactor in xylose metabolism and may alleviate the redox stress. The increased expression of genes involved in sulfur amino acid biosynthesis and the decreased expression of genes related to Fe(II) transport may alleviate redox stress as well. Meanwhile, it was remarkable that several glucose-repressible genes, including genes of the galactose metabolism, gluconeogenesis, and ethanol catabolism, had a lower expression level after adaptive evolution. Concomitantly, the expression levels of two regulators of the glucose signaling pathway, Rgs2 and Sip4, decreased, indicating a reshaped signaling pathway to xylose after adaptive evolution. Our findings provide new targets for construction of a superior bioethanol producing strain through inverse metabolic engineering.






Similar content being viewed by others
References
Attfield PV, Bell PJ (2006) Use of population genetics to derive nonrecombinant Saccharomyces cerevisiae strains that grow using xylose as a sole carbon source. FEMS Yeast Res 6:862–868
Batt CA, Carvallo S, Easson DD, Akedo M, Sinskey AJ (1986) Direct evidence for a xylose metabolic pathway in Saccharomyces cerevisiae. Biotechnol Bioeng 28:549–553
Bergdahl B, Sandstrom AG, Borgstrom C, Boonyawan T, van Niel EW, Gorwa-Grauslund MF (2013) Engineering yeast hexokinase 2 for improved tolerance toward xylose-induced inactivation. PLoS One 8:e75055
Boer VM, de Winde JH, Pronk JT, Piper MD (2003) The genome-wide transcriptional responses of Saccharomyces cerevisiae grown on glucose in aerobic chemostat cultures limited for carbon, nitrogen, phosphorus, or sulfur. J Biol Chem 278:3265–3274
Çakar ZP, Alkım C, Turanlı B, Tokman N, Akman S, Sarıkaya M, Tamerler C, Benbadis L, François JM (2009) Isolation of cobalt hyper-resistant mutants of Saccharomyces cerevisiae by in vivo evolutionary engineering approach. J Biotechnol 143(2):130–138
Çakar ZP, Turanlı-Yıldız B, Alkım C, Yılmaz Ü (2012) Evolutionary engineering of Saccharomyces cerevisiae for improved industrially important properties. FEMS Yeast Res 12:171–182
Cardoso LA, Ferreira ST, Hermes-Lima M (2008) Reductive inactivation of yeast glutathione reductase by Fe(II) and NADPH. Comp Biochem Physiol A: Comp Physiol 151:313–321
Caro AA, Cederbaum AI (2004) Antioxidant properties of S-adenosyl-l-methionine in Fe2+-initiated oxidations. Free Radic Biol Med 36:1303–1316
Chu BC, Lee H (2007) Genetic improvement of Saccharomyces cerevisiae for xylose fermentation. Biotechnol Adv 25:425–441
Demeke MM, Dietz H, Li Y, Foulquié-Moreno MR, Mutturi S, Deprez S, Den Abt T, Bonini BM, Liden G, Dumortier F, Verplaetse A, Boles E, Thevelein JM (2013) Development of a D-xylose fermenting and inhibitor tolerant industrial Saccharomyces cerevisiae strain with high performance in lignocellulose hydrolysates using metabolic and evolutionary engineering. Biotechnol Biofuels 6:89
Dong YX, Sueda S, Nikawa JI, Kondo H (2004) Characterization of the products of the genes SNO1 and SNZ1 involved in pyridoxine synthesis in Saccharomyces cerevisiae. Eur J Biochem 271:745–752
Dragosits M, Mattanovich D (2013) Adaptive laboratory evolution—principles and applications for biotechnology. Microb Cell Factories 12:1
Drake JW (1991) A constant rate of spontaneous mutation in DNA-based microbes. Proc Natl Acad Sci U S A 88:7160–7164
Du J, Yuan Y, Si T, Lian J, Zhao H (2012) Customized optimization of metabolic pathways by combinatorial transcriptional engineering. Nucleic Acids Res 40:e142–e142
Elena SF, Lenski RE (2003) Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat Rev Genet 4:457–469
Gancedo JM (1998) Yeast carbon catabolite repression. Microbiol Mol Biol Rev 62:334–361
Greenall A, Lei G, Swan DC, James K, Wang L, Peters H, Wipat A, Wilkinson DJ, Lydall D (2008) A genome wide analysis of the response to uncapped telomeres in budding yeast reveals a novel role for the NAD+ biosynthetic gene BNA2 in chromosome end protection. Genome Biol 9:R146
Hohmann S, Meacock PA (1998) Thiamin metabolism and thiamin diphosphate-dependent enzymes in the yeast Saccharomyces cerevisiae: genetic regulation. Biochim Biophys Acta 1385:201–219
Jeppsson M, Johansson B, Hahn-Hägerdal B, Gorwa-Grauslund MF (2002) Reduced oxidative pentose phosphate pathway flux in recombinant xylose-utilizing Saccharomyces cerevisiae strains improves the ethanol yield from xylose. Appl Environ Microbiol 68:1604–1609
Jin YS, Ni H, Laplaza JM, Jeffries TW (2003) Optimal growth and ethanol production from xylose by recombinant Saccharomyces cerevisiae require moderate D-xylulokinase activity. Appl Environ Microbiol 69:495–503
Jin YS, Laplaza JM, Jeffries TW (2004) Saccharomyces cerevisiae engineered for xylose metabolism exhibits a respiratory response. Appl Environ Microbiol 70:6816–6825
Kida K, Kume K, Morimura S, Sonoda Y (1992) Repeated-batch fermentation process using a thermotolerant flocculating yeast constructed by protoplast fusion. J Fermentation Bioeng 74:169–173
Kim JM, Vanguri S, Boeke JD, Gabriel A, Voytas DF (1998) Transposable elements and genome organization: a comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res 8:464–478
Krahulec S, Klimacek M, Nidetzky B (2012) Analysis and prediction of the physiological effects of altered coenzyme specificity in xylose reductase and xylitol dehydrogenase during xylose fermentation by Saccharomyces cerevisiae. J Biotechnol 158:192–202
Kuttykrishnan S, Sabina J, Langton LL, Johnston M, Brent MR (2010) A quantitative model of glucose signaling in yeast reveals an incoherent feed forward loop leading to a specific, transient pulse of transcription. Proc Natl Acad Sci U S A 107:16743–16748
Kuyper M, Aaron A, van Dijken JP, Pronk JT (2004) Minimal metabolic engineering of Saccharomyces cerevisiae for efficient anaerobic xylose fermentation: a proof of principle. FEMS Yeast Res 4:655–664
Kuyper M, Toirkens MJ, Diderich JA, Winkler AA, van Dijken JP, Pronk JT (2005) Evolutionary engineering of mixed-sugar utilization by a xylose-fermenting Saccharomyces cerevisiae strain. FEMS Yeast Res 5:925–934
Latimer LN, Lee ME, Medina-Cleghorn D, Kohnz RA, Nomura DK, Dueber JE (2014) Employing a combinatorial expression approach to characterize xylose utilization in Saccharomyces cerevisiae. Metab Eng 25:20–29
Lee SM, Jellison T, Alper HS (2014) Systematic and evolutionary engineering of a xylose isomerase-based pathway in Saccharomyces cerevisiae for efficient conversion yields. Biotechnol Biofuels 7:1
Lee SY, Kim HU (2015) Systems strategies for developing industrial microbial strains. Nature Biotechnol 33:1061–1072
Li YC, Mitsumasu K, Gou ZX, Gou M, Tang YQ, Li GY, XL W, Akamatsu T, Taguchi H, Kida K (2015) Xylose fermentation efficiency and inhibitor tolerance of the recombinant industrial Saccharomyces cerevisiae strain NAPX37. Appl Microbiol Biotechnol 100:1531–1542
Liu E, Hu Y (2010) Construction of a xylose-fermenting Saccharomyces cerevisiae strain by combined approaches of genetic engineering, chemical mutagenesis and evolutionary adaptation. Biochem Eng J 48:204–210
Lu C, Jeffries T (2007) Shuffling of promoters for multiple genes to optimize xylose fermentation in an engineered Saccharomyces cerevisiae strain. Appl Environ Microbiol 73:6072–6077
Lu SC (2000) S-adenosylmethionine. Int J Biochem Cell Biol 32:391–395
Matsushika A, Inoue H, Kodaki T, Sawayama S (2009) Ethanol production from xylose in engineered Saccharomyces cerevisiae strains, current state and perspectives. Appl Microbiol Biotechnol 84(1):37–53
Matsushika A, Goshima T, Hoshino T (2014) Transcription analysis of recombinant industrial and laboratory Saccharomyces cerevisiae strains reveals the molecular basis for fermentation of glucose and xylose. Microb Cell Factories 13:459–459
Mojzita D, Hohmann S (2006) Pdc2 coordinates expression of the THI regulon in the yeast Saccharomyces cerevisiae. Mol Gen Genomics 276:147–161
Oud B, van Maris AJ, Daran JM, Pronk JT (2012) Genome-wide analytical approaches for reverse metabolic engineering of industrially relevant phenotypes in yeast. FEMS Yeast Res 12:183–196
Pannunzio NR, Manthey GM, Liddell LC, BX F, Roberts CM, Bailis AM (2012) Rad59 regulates association of Rad52 with DNA double-strand breaks. Microbiology 1:285–297
Patterson MN, Scannapieco AE, PH A, Dorsey S, Royer CA, Maxwell PH (2015) Preferential retrotransposition in aging yeast mother cells is correlated with increased genome instability. DNA Repair 34:18–27
Peng B, Shen Y, Li X, Chen X, Hou J, Bao X (2012) Improvement of xylose fermentation in respiratory-deficient xylose-fermenting Saccharomyces cerevisiae. Metab Eng 14:9–18
Pittet M, Conzelmann A (2007) Biosynthesis and function of GPI proteins in the yeast Saccharomyces cerevisiae. Biochim Biophys Acta 1771:405–420
Pollak N, Dölle C, Ziegler M (2007) The power to reduce: pyridine nucleotides--small molecules with a multitude of functions. Biochem J 402:205–218
Rapala-Kozik M, Kowalska E, Ostrowska K (2008) Modulation of thiamine metabolism in Zea mays seedlings. J Exp Bot 59:4133–4143
Rapala-Kozik M, Wolak N, Kujda M, Banas AK (2012) The upregulation of thiamine (vitamin B 1) biosynthesis in Arabidopsis thaliana seedlings under salt and osmotic stress conditions is mediated by abscisic acid at the early stages of this stress response. BMC Plant Biol 12:1
Regenberg B, Regenberg B, Grotkjær T, Winther O, Fausbøll A, Åkesson M, Bro C, Hansen LK, Brunak S, Nielsen J (2006) Growth-rate regulated genes have profound impact on interpretation of transcriptome profiling in Saccharomyces cerevisiae. Genome Biol 7:R107
Robberecht C, Voet T, Esteki MZ, Nowakowska BA, Vermeesch JR (2012) Nonallelic homologous recombination between retrotransposable elements is a driver of de novo unbalanced translocations. Genome Res 23:411–418
Rodriguez-Navarro S, Llorente B, Rodriguez-Manzaneque MT, Ramne A, Uber G, Marchesan D, Dujon B, Herrero E, Sunnerhagen P, Perez-Ortin JE (2002) Functional analysis of yeast gene families involved in metabolism of vitamins B1 and B6. Yeast 19:1261–1276
Rodriguez A, Herrero P, Moreno F (2001) The hexokinase 2 protein regulates the expression of the GLK1, HXK1 and HXK2 genes of Saccharomyces cerevisiae. Biochem J 355:625–631
Runquist D, Hahn-Hägerdal B, Bettiga M (2009) Increased expression of the oxidative pentose phosphate pathway and gluconeogenesis in anaerobically growing xylose-utilizing Saccharomyces cerevisiae. Microb Cell Factories 8:49
Salusjärvi L, Kankainen M, Soliymani R, Pitkänen JP, Penttilä M, Ruohonen L (2008) Regulation of xylose metabolism in recombinant Saccharomyces cerevisiae. Microb Cell Factories 7:18
Shen Y, Chen X, Peng B, Chen L, Jin H, Bao X (2012) An efficient xylose-fermenting recombinant Saccharomyces cerevisiae strain obtained through adaptive evolution and its global transcription profile. Appl Microbiol Biotechnol 96:1079–1091
Shen Y, Hou J, Bao X (2013) Enhanced xylose fermentation capacity related to an altered glucose sensing and repression network in a recombinant Saccharomyces cerevisiae. Bioengineered 4:435–437
Sims RE, Mabee W, Saddler JN, Taylor M (2010) An overview of second generation biofuel technologies. Bioresour Technol 101:1570–1580
Sonderegger M, Sauer U (2003) Evolutionary engineering of Saccharomyces cerevisiae for anaerobic growth on xylose. Appl Environ Microbiol 69:1990–1998
Taddei F, Radman M, Maynard-Smith J, Toupance B, Gouyon P, Godelle B (1997) Role of mutator alleles in adaptive evolution. Nature 387:700–702
Tang Y, An M, Liu K, Nagai S, Shigematsu T, Morimura S, Kida K (2006) Ethanol production from acid hydrolysate of wood biomass using the flocculating yeast Saccharomyces cerevisiae strain KF-7. Proc Biochem 41:909–914
Tanghe A, Prior B, Thevelein JM (2006) Yeast responses to stresses. In: Rosa CA, Peter G (eds) Biodiversity and ecophysiology of yeasts. Springer-Verlag, Berlin, pp 175–195
Thomas D, SurdinKerjan Y (1997) Metabolism of sulfur amino acids in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 61:503–532
Wahlbom CF, Cordero Otero RR, van Zyl WH, Hahn-Hägerdal B, Jonsson LJ (2003) Molecular analysis of a Saccharomyces cerevisiae mutant with improved ability to utilize xylose shows enhanced expression of proteins involved in transport, initial xylose metabolism, and the pentose phosphate pathway. Appl Environ Microbiol 69:740–746
Watanabe S, Abu Saleh A, Pack SP, Annaluru N, Kodaki T, Makino K (2007) Ethanol production from xylose by recombinant Saccharomyces cerevisiae expressing protein-engineered NADH-preferring xylose reductase from Pichia stipitis. Microbiology 153:3044–3054
Wolak N, Kowalska E, Kozik A, Rapala-Kozik M (2014) Thiamine increases the resistance of baker's yeast Saccharomyces cerevisiae against oxidative, osmotic and thermal stress, through mechanisms partly independent of thiamine diphosphate-bound enzymes. FEMS Yeast Res 14:1249–1262
Yun CW (2000) Siderophore-iron uptake in Saccharomyces cerevisiae. J Biol Chem 275:16354–16359
Zaman S, Lippman SI, Zhao X, Broach JR (2008) How Saccharomyces responds to nutrients. Annu Rev Genet 42:27–81
Zeng WY, Tang YQ, Gou M, Xia ZY, Kida K (2016) Transcriptomes of a xylose-utilizing industrial flocculating Saccharomyces cerevisiae strain cultured in media containing different sugar sources. AMB Express 6:51
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31170093).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare no conflict of interest.
Electronic supplementary material
ESM 1
(PDF 918 kb)
Rights and permissions
About this article
Cite this article
Zeng, WY., Tang, YQ., Gou, M. et al. Comparative transcriptomes reveal novel evolutionary strategies adopted by Saccharomyces cerevisiae with improved xylose utilization capability. Appl Microbiol Biotechnol 101, 1753–1767 (2017). https://doi.org/10.1007/s00253-016-8046-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-016-8046-y