Abstract
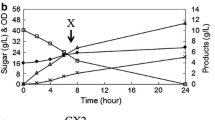
A heterologous xylose utilization pathway, either xylose reductase–xylitol dehydrogenase (XR–XDH) or xylose isomerase (XI), is usually introduced into Saccharomyces cerevisiae to construct a xylose-fermenting strain for lignocellulosic ethanol production. To investigate the molecular basis underlying the effect of different xylose utilization pathways on the xylose metabolism and ethanol fermentation, transcriptomes of flocculating industrial strains with the same genetic background harboring different xylose utilization pathways were studied. A different source of xylA did not obviously affect the change of the strains transcriptome, but compared with the XR–XDH strain, several key genes in the central carbon pathway were downregulated in the XI strains, suggesting a lower carbon flow to ethanol. The carbon starvation caused by lower xylose metabolism in XI strains further influenced the stress response and cell metabolism of amino acid, nucleobase, and vitamin. Besides, the downregulated genes mostly included those involved in mitotic cell cycle and the cell division–related process. Moreover, the transcriptomes analysis indicated that the after integrate xylA in the δ region, the DNA and chromosome stability and cell wall integrity of the strains were affected to some extent. The aim of this was to provide some reference for constructing efficient xylose-fermenting strains.





Similar content being viewed by others
References
Zabed, H., Sahu, J. N., Boyce, A. N., & Faruq, G. (2016). Fuel ethanol production from lignocellulosic biomass: an overview on feedstocks and technological approaches. Renewable and Sustainable Energy Reviews, 66, 751–774.
Hoang, P. T. N., Ko, J. K., Gong, G., Um, Y., & Lee, S. M. (2018). Genomic and phenotypic characterization of a refactored xylose-utilizing Saccharomyces cerevisiae strain for lignocellulosic biofuel production. Biotechnology for Biofuels, 11(1), 268.
Cheng, C., Tang, R. Q., Xiong, L., Hector, R. E., Bai, F. W., & Zhao, X. Q. (2018). Association of improved oxidative stress tolerance and alleviation of glucose repression with superior xylose-utilization capability by a natural isolate of Saccharomyces cerevisiae. Biotechnology for Biofuels, 11(1), 28.
Cai, Z., Zhang, B., & Li, Y. (2012). Engineering Saccharomyces cerevisiae for efficient anaerobic xylose fermentation: reflections and perspectives. Biotechnology Journal, 7(1), 34–46.
Demeke, M. M., Dietz, H., Li, Y., Foulquié-Moreno, M. R., Mutturi, S., Deprez, S., Den Abt, T., Bonini, B. M., Liden, G., Dumortier, F., Verplaetse, A., Boles, E., & Thevelein, J. M. (2013). Development of a D-xylose fermenting and inhibitor tolerant industrial Saccharomyces cerevisiae strain with high performance in lignocellulose hydrolysates using metabolic and evolutionary engineering. Biotechnology for Biofuels, 6(1), 89.
Bergdahl, B., Heer, D., Sauer, U., Hahn-Hägerdal, B., & van Niel, E. W. (2012). Dynamic metabolomics differentiates between carbon and energy starvation in recombinant Saccharomyces cerevisiae fermenting xylose. Biotechnology for Biofuels, 5(1), 34.
Li, Y.-C., Mitsumasu, K., Gou, Z.-X., Gou, M., Tang, Y.-Q., Li, G. Y., Wu, X. L., Akamatsu, T., Taguchi, H., & Kida, K. (2016). Xylose fermentation efficiency and inhibitor tolerance of the recombinant industrial Saccharomyces cerevisiae strain NAPX37. Applied Microbiology and Biotechnology, 100(3), 1531–1542.
Li, Y.-C., Li, G.-Y., Gou, M., Xia, Z. Y., Tang, Y.-Q., & Kida, K. (2016). Functional expression of xylose isomerase in flocculating industrial Saccharomyces cerevisiae strain for bioethanol production. Journal of Bioscience and Bioengineering, 121(6), 685–691.
Li, Y.-C., Zeng, W.-Y., Gou, M., Sun, Z. Y., Xia, Z.-Y., & Tang, Y.-Q. (2017). Transcriptome changes in adaptive evolution of xylose-fermenting industrial Saccharomyces cerevisiae strains with δ-integration of different xyla genes. Applied Microbiology and Biotechnology, 101, 1–13.
Wang, G., Tan, L., Sun, Z. Y., Gou, Z. X., Tang, Y. Q., & Kida, K. (2014). Production of bioethanol from rice straw by simultaneous saccharification and fermentation of whole pretreated slurry using Saccharomyces cerevisiae KF-7. Environmental Progress and Sustainable Energy, 34, 582–588.
Tang, Y., An, M., Liu, K., Nagai, S., Shigematsu, T., Morimura, S., & Kida, K. (2006). Ethanol production from acid hydrolysate of wood biomass using the flocculating yeast Saccharomyces cerevisiae strain KF-7. Process Biochemistry, 41(4), 909–914.
Gao, C., Fu, Q., Su, B., Zhou, S., Liu, F., Song, L., Zhang, M., Ren, Y., Dong, X., Tan, F., & Li, C. (2016). Transcriptomic profiling revealed the signatures of intestinal barrier alteration and pathogen entry in turbot (Scophthalmus maximus) following Vibrio anguillarum challenge. Developmental and Comparative Immunology, 65, 159–168.
Tan, S., & Richmond, T. J. (1998). Eukaryotic transcription factors. Current Opinion in Structural Biology, 8(1), 41–48.
Feng, X., & Zhao, H. (2013). Investigating host dependence of xylose utilization in recombinant Saccharomyces cerevisiae strains using RNA-Seq analysis. Biotechnology for Biofuels, 6(1), 96.
Cipollina, C., Alberghina, L., Porro, D., & Vai, M. (2005). SFP1 is involved in cell size modulation in respiro-fermentative growth conditions. Yeast, 22(5), 385–399.
King, L., & Butler, G. (1998). Ace2p, a regulator of CTS1 (chitinase) expression, affects pseudohyphal production in Saccharomyces cerevisiae. Current Genetics, 34(3), 183–191.
Schmitt, A. P., & Mcentee, K. (1996). Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences, 93(12), 5777–5782.
Cullen, P. J., & Sprague, G. F. (2000). Glucose depletion causes haploid invasive growth in yeast. PNAS, 97(25), 13619–13624.
Jin, Y. S., Laplaza, J. M., & Jeffries, T. W. (2004). Saccharomyces cerevisiae engineered for xylose metabolism exhibits a respiratory response. Applied and Environmental Microbiology, 70(11), 6816–6825.
Qun, Y., Runxiang, Q., Foland, T. B., Griesen, D., Gallowa, C. S., Chiu, Y. H., Sandmeier, J., Broach, J. R., & Bi, X. (2003). Rap1p and other transcriptional regulators can function in defining distinct domains of gene expression. Nucleic Acids Research, 31, 1224–1233.
Garcia-Gimeno, M. A., & Struhl, K. (2000). Aca1 and Aca2, ATF/CREB activators in Saccharomyces cerevisiae, are important for carbon source utilization but not the response to stress. Molecular and Cellular Biology, 20(12), 4340–4349.
Matsushika, A., Inoue, H., Kodaki, T., & Sawayama, S. (2009). Ethanol production from xylose in engineered Saccharomyces cerevisiae strains: current state and perspectives. Applied Microbiology and Biotechnology, 84(1), 37–53.
Matsushika, A., Goshima, T., & Hoshino, T. (2014). Transcription analysis of recombinant industrial and laboratory Saccharomyces cerevisiae strains reveals the molecular basis for fermentation of glucose and xylose. Microbial Cell Factories, 13(1), 16.
Sedlak, M., & Ho, N. W. Y. (2004). Characterization of the effectiveness of hexose transporters for transporting xylose during glucose and xylose co-fermentation by a recombinant Saccharomyces yeast. Yeast, 21(8), 671–684.
Zeng, W.-Y., Tang, Y.-Q., Gou, M., Xia, Z. Y., & Kida, K. (2016). Transcriptomes of a xylose-utilizing industrial flocculating Saccharomyces cerevisiae strain cultured in media containing different sugar sources. AMB Express, 6(1), 51.
Toivari, M. H., Aristidou, A., Ruohonen, L., & Penttilä, M. (2001). Conversion of xylose to ethanol by recombinant Saccharomyces cerevisiae: importance of xylulokinase (XKS1) and oxygen availability. Metabolic Engineering, 3(3), 236–249.
Ismail, K. S. K., Sakamoto, T., Hasunuma, T., & Kondo, A. (2013). Time-based comparative transcriptomics in engineered xylose-utilizing Saccharomyces cerevisiae identifies temperature-responsive genes during ethanol production. Journal of Industrial Microbiology and Biotechnology, 40(9), 1039–1050.
Kresnowati, M. T. A. P., van Winden, W. A., Almering, M. J. H., Ten Pierick, A., Ras, C., Knijnenburg, T. A., Daran-Lapujade, P., Pronk, J. T., Heijnen, J. J., & Daran, J. M. (2006). When transcriptome meets metabolome: Fast cellular responses of yeast to sudden relief of glucose limitation. Molecular Systems Biology, 2, 16.
Zeng, W.-Y., Tang, Y.-Q., Gou, M., Sun, Z. Y., Xia, Z. Y., & Kida, K. (2017). Comparative transcriptomes reveal novel evolutionary strategies adopted by Saccharomyces cerevisiae with improved xylose utilization capability. Applied Microbiology and Biotechnology, 101(4), 1753–1767.
Piper, M. D., Hong, S. P., Ball, G. E., & Dawes, I. W. (2000). Regulation of the balance of one-carbon metabolism in Saccharomyces cerevisiae. The Journal of Biological Chemistry, 275(40), 30987–30995.
Frýdlová, I., Malcová, I., Vašicová, P., & Hašek, J. (2009). Deregulation of DSE1 gene expression results in aberrant budding within the birth scar and cell wall integrity pathway activation in Saccharomyces cerevisiae. Eukaryotic Cell, 8(4), 586–594.
Janik, A., Sosnowska, M., Kruszewska, J., Krotkiewski, H., Lehle, L., & Palamarczyk, G. (2003). Overexpression of GDP-mannose pyrophosphorylase in Saccharomyces cerevisiae corrects defects in dolichol-linked saccharide formation and protein glycosylation. Biochimica et Biophysica Acta, 1621(1), 22–30.
Krysan, D. J., Ting, E. L., Abeijon, C., Kroos, L., & Fuller, R. S. (2005). Yapsins are a family of aspartyl proteases required for cell wall integrity in Saccharomyces cerevisiae. Eukaryotic Cell, 4(8), 1364–1374.
Sumita, T., Yoko-o, T., Shimma, Y., & Jigami, Y. (2005). Comparison of cell wall localization among pir family proteins and functional dissection of the region required for cell wall binding and bud scar recruitment of Pir1p. Eukaryotic Cell, 4(11), 1872–1881.
Klis, F. M., Boorsma, A., & De Groot, P. W. (2006). Cell wall construction in Saccharomyces cerevisiae. Yeast, 23(3), 185–202.
de Lucena, R. M., Elsztein, C., de Barros Pita, W., de Souza, R. B., de Sá Leitão Paiva Júnior, S., & de Morais Junior, M. A. (2015). Transcriptomic response of Saccharomyces cerevisiae for its adaptation to sulphuric acid-induced stress. Antonie Van Leeuwenhoek, 108(5), 1147–1160.
Funding
This work was supported by the National Natural Science Foundation of China (31170093), the Talent Project for Science and Technology Innovation of Sichuan Province (2017RZ0021), and the Open Fund of Key Laboratory of Development and Application of Rural Renewable Energy, Ministry of Agriculture of China (2018-009).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Li, YC., Xie, CY., Yang, BX. et al. Comparative Transcriptome Analysis of Recombinant Industrial Saccharomyces cerevisiae Strains with Different Xylose Utilization Pathways. Appl Biochem Biotechnol 189, 1007–1019 (2019). https://doi.org/10.1007/s12010-019-03060-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-019-03060-8