Abstract
The cell envelope is an essential bacterial structure that consists of the cytoplasmic membrane, the cell wall, and—in Gram-negative bacteria—the outer membrane. Because of its crucial functions, it represents a prime antibiotic target. Monitoring and maintaining its integrity are therefore keys to survival, especially in competitive environments where antibiotics represent one means of suppressing the growth of competitors. Resistance against external antibiotic threat, as well as auto-immunity against self-produced antibiotics, is often mediated by two-component systems (2CSs). They respond to antibiotic threat by inducing gene expression that results in the production of specific resistance determinants. The underlying transcriptional control is exhibited at the level of specific target promoters, which usually share a number of relevant features: They are tightly controlled and only induced in the presence of specific (sets of) antibiotics. This induction is dose dependent and often very sensitive, that is, it occurs well below inhibitory antibiotic concentrations. Because of these characteristics, a number of well-characterized cell envelope stress-inducible promoters have been developed for two different applied purposes: first, as whole-cell biosensors for antibiotic detection and mechanism-of-action studies, and second, as antibiotic-inducible expression systems for biotechnological purposes. The current state of research in both fields will be discussed in this review, focusing on 2CS-regulated promoters from Firmicutes bacteria that are induced to mediate resistance against antimicrobial peptides (AMPs) targeting the cell envelope.


Similar content being viewed by others
References
Bandow JE, Brotz H, Leichert LI, Labischinski H, Hecker M (2003) Proteomic approach to understanding antibiotic action. Antimicrob Agents Chemother 47:948–955
Bassetti M, Merelli M, Temperoni C, Astilean A (2013) New antibiotics for bad bugs: where are we? Ann Clin Microbiol Antimicrob 12:22. doi:10.1186/1476-0711-12-22
Bernaudat F, Frelet-Barrand A, Pochon N, Dementin S, Hivin P, Boutigny S, Rioux JB, Salvi D, Seigneurin-Berny D, Richaud P, Joyard J, Pignol D, Sabaty M, Desnos T, Pebay-Peyroula E, Darrouzet E, Vernet T, Rolland N (2011) Heterologous expression of membrane proteins: choosing the appropriate host. PLoS ONE 6:e29191. doi:10.1371/journal.pone.0029191
Bongers RS, Veening J-W, Van Wieringen M, Kuipers OP, Kleerebezem M (2005) Development and characterization of a subtilin-regulated expression system in Bacillus subtilis: strict control of gene expression by addition of subtilin. Appl Environ Microbiol 71:8818–8824
Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J (2009) Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis 48:1–12. doi:10.1086/595011
Breukink E, de Kruijff B (1999) The lantibiotic nisin, a special case or not? Biochim Biophys Acta 1462:223–234
Burkard M, Entian KD, Stein T (2007) Development and application of a microtiter plate-based autoinduction bioassay for detection of the lantibiotic subtilin. J Microbiol Methods 70:179–185. doi:10.1016/j.mimet.2007.04.015
Burkard M, Stein T (2008) Microtiter plate bioassay to monitor the interference of antibiotics with the lipid II cycle essential for peptidoglycan biosynthesis. J Microbiol Methods 75:70–74
Chen J, Rosen BP (2014) Biosensors for inorganic and organic arsenicals. Biosensors 4:494–512. doi:10.3390/bios4040494
Cortes J (2014) Lantibiotics and similar peptides produced by and active on gram-positives: discovery, development and perspectives. In: Marinelli F, Genilloud O (eds) Antimicrobials. Springer, Berlin-Heidelberg. doi:10.1007/978-3-642-39968-8_7
Czarny TL, Perri AL, French S, Brown ED (2014) Discovery of novel cell wall-active compounds using P ywaC , a sensitive reporter of cell wall stress, in the model gram-positive bacterium Bacillus subtilis. Antimicrob Agents Chemother 58:3261–3269. doi:10.1128/AAC.02352-14
D'Elia MA, Millar KE, Bhavsar AP, Tomljenovic AM, Hutter B, Schaab C, Moreno-Hagelsieb G, Brown ED (2009) Probing teichoic acid genetics with bioactive molecules reveals new interactions among diverse processes in bacterial cell wall biogenesis. Chem Biol 16:548–556. doi:10.1016/j.chembiol.2009.04.009
de Ruyter PG, Kuipers OP, de Vos WM (1996) Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl Environ Microbiol 62:3662–3667
Delves-Broughton J, Blackburn P, Evans RJ, Hugenholtz J (1996) Applications of the bacteriocin, nisin. Antonie Van Leeuwenhoek 69:193–202
Dintner S, Staron A, Berchtold E, Petri T, Mascher T, Gebhard S (2011) Co-evolution of ABC-transporters and two-component regulatory systems as resistance modules against antimicrobial peptides in Firmicutes bacteria. J Bacteriol 193:3851–3862. doi:10.1128/JB.05175-11
Eiamphungporn W, Helmann JD (2008) The Bacillus subtilis σM regulon and its contribution to cell envelope stress responses. Mol Microbiol 67:830–848
Eichenbaum Z, Federle MJ, Marra D, de Vos WM, Kuipers OP, Kleerebezem M, Scott JR (1998) Use of the lactococcal nisA promoter to regulate gene expression in gram-positive bacteria: comparison of induction level and promoter strength. Appl Environ Microbiol 64:2763–2769
Fantino JR, Barras F, Denizot F (2009) Sposensor: a whole-bacterial biosensor that uses immobilized Bacillus subtilis spores and a one-step incubation/detection process. J Mol Microbiol Biotechnol 17:90–95. doi:10.1159/000206634
Fernandez-Lopez R, Ruiz R, de la Cruz F, Moncalian G (2015) Transcription factor-based biosensors enlightened by the analyte. Front Microbiol 6:648. doi:10.3389/fmicb.2015.00648
Fischbach MA, Walsh CT (2009) Antibiotics for emerging pathogens. Science 325:1089–1093. doi:10.1126/science.1176667
Fritz G, Dintner S, Treichel NS, Radeck J, Gerland U, Mascher T, Gebhard S (2015) A new way of sensing: need-based activation of antibiotic resistance by a flux-sensing mechanism. MBio 6. doi:10.1128/mBio.00975-15
Gaspar P, Carvalho AL, Vinga S, Santos H, Neves AR (2013) From physiology to systems metabolic engineering for the production of biochemicals by lactic acid bacteria. Biotechnol Adv 31:764–788. doi:10.1016/j.biotechadv.2013.03.011
Gebhard S (2012) ABC transporters of antimicrobial peptides in Firmicutes bacteria—phylogeny, function and regulation. Mol Microbiol 86:1295–1317. doi:10.1111/mmi.12078
Hasper HE, de Kruijff B, Breukink E (2004) Assembly and stability of nisin-lipid II pores. Biochemistry 43:11567–11575. doi:10.1021/bi049476b
He W, Yuan S, Zhong WH, Siddikee MA, Dai CC (2016) Application of genetically engineered microbial whole-cell biosensors for combined chemosensing. Appl Microbiol Biotechnol 100:1109–1119. doi:10.1007/s00253-015-7160-6
Helmann JD, Mascher T (2007) Compositions and methods for screening antibacterial compounds. USA Patent US 07309484
Hutter B, Fischer C, Jacobi A, Schaab C, Loferer H (2004a) Panel of Bacillus subtilis reporter strains indicative of various modes of action. Antimicrob Agents Chemother 48:2588–2594
Hutter B, Schaab C, Albrecht S, Borgmann M, Brunner NA, Freiberg C, Ziegelbauer K, Rock CO, Ivanov I, Loferer H (2004b) Prediction of mechanisms of action of antibacterial compounds by gene expression profiling. Antimicrob Agents Chemother 48:2838–2844
Jordan S, Hutchings MI, Mascher T (2008) Cell envelope stress response in Gram-positive bacteria. FEMS Microbiol Rev 32:107–146
Jordan S, Junker A, Helmann JD, Mascher T (2006) Regulation of LiaRS-dependent gene expression in Bacillus subtilis: identification of inhibitor proteins, regulator binding sites and target genes of a conserved cell envelope stress-sensing two-component system. J Bacteriol 188:5153–5166
Jordan S, Rietkotter E, Strauch MA, Kalamorz F, Butcher BG, Helmann JD, Mascher T (2007) LiaRS-dependent gene expression is embedded in transition state regulation in Bacillus subtilis. Microbiology 153:2530–2540. doi:10.1099/mic.0.2007/006817-0
Juhas M, Reuss DR, Zhu B, Commichau FM (2014) Bacillus subtilis and Escherichia coli essential genes and minimal cell factories after one decade of genome engineering. Microbiology 160:2341–2351. doi:10.1099/mic.0.079376-0
Kleerebezem M (2004) Quorum sensing control of lantibiotic production; nisin and subtilin autoregulate their own biosynthesis. Peptides 25:1405–1414
Kleerebezem M, Bongers R, Rutten G, de Vos WM, Kuipers OP (2004) Autoregulation of subtilin biosynthesis in Bacillus subtilis: the role of the spa-box in subtilin-responsive promoters. Peptides 25:1415–1424. doi:10.1016/j.peptides.2003.11.025
Kobras CM, Mascher T, Gebhard S (2016) Application of a Bacillus subtilis whole-cell biosensor (P liaI -lux) for the identification of cell wall active antibacterial compounds. In: Methods in molecular biology. p in press
Kuipers OP, Beerthuyzen MM, de Ruyter PG, Luesink EJ, de Vos WM (1995) Autoregulation of nisin biosynthesis in Lactococcus lactis by signal transduction. J Biol Chem 270:27299–27304
Kuipers OP, Beerthuyzen MM, Siezen RJ, De Vos WM (1993) Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis. Requirement of expression of the nisA and nisI genes for development of immunity. Eur J Biochem FEBS 216:281–291
Liu L, Liu Y, Shin HD, Chen RR, Wang NS, Li J, Du G, Chen J (2013) Developing Bacillus spp. as a cell factory for production of microbial enzymes and industrially important biochemicals in the context of systems and synthetic biology. Appl Microbiol Biotechnol 97:6113–6127. doi:10.1007/s00253-013-4960-4
Maischberger T, Mierau I, Peterbauer CK, Hugenholtz J, Haltrich D (2010) High-level expression of Lactobacillus beta-galactosidases in Lactococcus lactis using the food-grade, nisin-controlled expression system NICE. J Agric Food Chem 58:2279–2287. doi:10.1021/jf902895g
Marciniak BC, Trip H, van-der Veek PJ, Kuipers OP (2012) Comparative transcriptional analysis of Bacillus subtilis cells overproducing either secreted proteins, lipoproteins or membrane proteins. Microb Cell Factories 11:66. doi:10.1186/1475-2859-11-66
Mascher T, Margulis NG, Wang T, Ye RW, Helmann JD (2003) Cell wall stress responses in Bacillus subtilis: the regulatory network of the bacitracin stimulon. Mol Microbiol 50:1591–1604
Mascher T, Zimmer SL, Smith TA, Helmann JD (2004) Antibiotic-inducible promoter regulated by the cell envelope stress-sensing two-component system LiaRS of Bacillus subtilis. Antimicrob Agents Chemother 48:2888–2896
Mierau I, Kleerebezem M (2005) 10 years of the nisin-controlled gene expression system (NICE) in Lactococcus lactis. Appl Microbiol Biotechnol 68:705–717. doi:10.1007/s00253-005-0107-6
Mierau I, Leij P, van Swam I, Blommestein B, Floris E, Mond J, Smid EJ (2005a) Industrial-scale production and purification of a heterologous protein in Lactococcus lactis using the nisin-controlled gene expression system NICE: the case of lysostaphin. Microb Cell Factories 4:15. doi:10.1186/1475-2859-4-15
Mierau I, Olieman K, Mond J, Smid EJ (2005b) Optimization of the Lactococcus lactis nisin-controlled gene expression system NICE for industrial applications. Microb Cell Factories 4:16. doi:10.1186/1475-2859-4-16
Miller JH (1972) Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York
Mironczuk AM, Krasowska A, Murzyn A, Plachetka M, Lukaszewicz M (2012) Production of the Bacillus licheniformis SubC protease using Lactococcus lactis NICE expression system. Springer Plus 1:54. doi:10.1186/2193-1801-1-54
Morello E, Bermudez-Humaran LG, Llull D, Sole V, Miraglio N, Langella P, Poquet I (2008) Lactococcus lactis, an efficient cell factory for recombinant protein production and secretion. J Mol Microbiol Biotechnol 14:48–58. doi:10.1159/000106082
Ohki R, Giyanto TK, Masuyama W, Moriya S, Kobayashi K, Ogasawara N (2003) The BceRS two-component regulatory system induces expression of the bacitracin transporter, BceAB, in Bacillus subtilis. Mol Microbiol 49:1135–1144
Park M, Tsai SL, Chen W (2013) Microbial biosensors: engineered microorganisms as the sensing machinery. Sensors (Basel) 13:5777–5795. doi:10.3390/s130505777
Pendleton JN, Gorman SP, Gilmore BF (2013) Clinical relevance of the ESKAPE pathogens. Expert Rev Anti-Infect Ther 11:297–308. doi:10.1586/eri.13.12
Pinto D, Mascher T (2016) (Actino) Bacterial “intelligence”: using comparative genomics to unravel the information processing capacities of microbes. doi:10.1007/s00294-016-0569-3. Current Genetics
Radeck J, Gebhard S, Orchard PS, Kirchner M, Bauer S, Mascher T, Fritz G (2016) Anatomy of the bacitracin resistance network in Bacillus subtilis. Mol Microbiol. doi:10.1111/mmi.13336
Radeck J, Kraft K, Bartels J, Cikovic T, Dürr F, Emenegger J, Kelterborn S, Sauer C, Fritz G, Gebhard S, Mascher T (2013) The Bacillus BioBrick Box: generation and evaluation of essential genetic building blocks for standardized work with Bacillus subtilis. J Biol Eng 7:29. doi:10.1186/1754-1611-7-29
Raivio TL (2005) Envelope stress responses and Gram-negative bacterial pathogenesis. Mol Microbiol 56:1119–1128
Revilla-Guarinos A, Gebhard S, Mascher T, Zuniga M (2014) Defence against antimicrobial peptides: different strategies in Firmicutes. Environ Microbiol. doi:10.1111/1462-2920.12400
Rietkötter E, Hoyer D, Mascher T (2008) Bacitracin sensing in Bacillus subtilis. Mol Microbiol 68:768–785
Ron EZ (2007) Biosensing environmental pollution. Curr Opin Biotechnol 18:252–256. doi:10.1016/j.copbio.2007.05.005
Salzberg LI, Luo Y, Hachmann AB, Mascher T, Helmann JD (2011) The Bacillus subtilis GntR family repressor YtrA responds to cell wall antibiotics. J Bacteriol 193(20):5793–5801
Schallmey M, Singh A, Ward OP (2004) Developments in the use of Bacillus species for industrial production. Can J Microbiol 50:1–17
Schneider T, Kruse T, Wimmer R, Wiedemann I, Sass V, Pag U, Jansen A, Nielsen AK, Mygind PH, Raventos DS, Neve S, Ravn B, Bonvin AMJJ, De Maria L, Andersen AS, Gammelgaard LK, Sahl H-G, Kristensen H-H (2010) Plectasin, a fungal defensin, targets the bacterial cell wall precursor lipid II. Science 328:1168–1172. doi:10.1126/science.1185723
Schrecke K, Staroń A, Mascher T (2012) Two-component signaling in the Gram-positive envelope stress response: intramembrane-sensing histidine kinases and accessory membrane proteins. In: Gross R, Beier D (eds) Two component systems in bacteria. Horizon Scientific Press, Hethersett, Norwich, UK, pp. 199–229
Siezen RJ, Kuipers OP, de Vos WM (1996) Comparison of lantibiotic gene clusters and encoded proteins. Antonie Van Leeuwenhoek 69:171–184
Staroń A, Finkeisen DE, Mascher T (2011) Peptide antibiotic sensing and detoxification modules of Bacillus subtilis. Antimicrob Agents Chemother 55:515–525
Stein T, Borchert S, Kiesau P, Heinzmann S, Kloss S, Klein C, Helfrich M, Entian K-D (2002) Dual control of subtilin biosynthesis and immunity in Bacillus subtilis. Mol Microbiol 44:403–416. doi:10.1046/j.1365-2958.2002.02869.x
Stein T, Heinzmann S, Kiesau P, Himmel B, Entian KD (2003) The spa-box for transcriptional activation of subtilin biosynthesis and immunity in Bacillus subtilis. Mol Microbiol 47:1627–1636
Toymentseva AA, Schrecke K, Sharipova MR, Mascher T (2012) The LIKE system, a novel protein expression toolbox for Bacillus subtilis based on the liaI promoter. Microb Cell Factories 11:143. doi:10.1186/1475-2859-11-143
Urban A, Eckermann S, Fast B, Metzger S, Gehling M, Ziegelbauer K, Rübsamen-Waigmann H, Freiberg C (2007) Novel whole-cell antibiotic biosensors for compound discovery. Appl Environ Microbiol 73:6436–6443. doi:10.1128/aem.00586-07
van Dijl JM, Hecker M (2013) Bacillus subtilis: from soil bacterium to super-secreting cell factory. Microb Cell Factories 12:3. doi:10.1186/1475-2859-12-3
Virolainen N, Karp M (2014) Biosensors, antibiotics and food. Adv Biochem Eng Biotechnol 145:153–185. doi:10.1007/978-3-662-43619-6_5
Waidmann MS, Bleichrodt FS, Laslo T, Riedel CU (2011) Bacterial luciferase reporters: the Swiss army knife of molecular biology. Bioeng Bugs 2:8–16. doi:10.4161/bbug.2.1.13566
Walsh CT, Wencewicz TA (2014) Prospects for new antibiotics: a molecule-centered perspective. J Antibiot 67:7–22. doi:10.1038/ja.2013.49
Wecke T, Bauer T, Harth H, Mäder U, Mascher T (2011) The rhamnolipid stress response of Bacillus subtilis. FEMS Microbiol Lett 323(2):113–23. doi:10.1111/j.1574-6968.2011.02367.x
Wecke T, Mascher T (2011) Antibiotic research in the age of omics: from expression profiles to interspecies communication. J Antimicrobial Chemother 66:2689–2704. doi:10.1093/jac/dkr373
Wecke T, Zühlke D, Mäder U, Jordan S, Voigt B, Pelzer S, Labischinski H, Homuth G, Hecker M, Mascher T (2009) Daptomycin versus friulimicin B: in-depth profiling of Bacillus subtilis cell envelope stress responses. Antimicrob Agents Chemother 53:1619–1623. doi:10.1128/aac.01046-08
Wenzel M, Chiriac AI, Otto A, Zweytick D, May C, Schumacher C, Gust R, Albada HB, Penkova M, Kramer U, Erdmann R, Metzler-Nolte N, Straus SK, Bremer E, Becher D, Brotz-Oesterhelt H, Sahl HG, Bandow JE (2014) Small cationic antimicrobial peptides delocalize peripheral membrane proteins. Proc Natl Acad Sci U S A 111:E1409–E1418. doi:10.1073/pnas.1319900111
WHO (2014) Antimicrobial resistance: global report on surveillance.
Wiegert T, Homuth G, Versteeg S, Schumann W (2001) Alkaline shock induces the Bacillus subtilis σW regulon. Mol Microbiol 41:59–71
Wilson T, Hastings JW (1998) Bioluminescence. Annu Rev Cell Dev Biol 14:197–230. doi:10.1146/annurev.cellbio.14.1.197
Wolf D, Dominguez-Cuevas P, Daniel RA, Mascher T (2012) Cell envelope stress response in cell wall-deficient L-forms of Bacillus subtilis. Antimicrob Agents Chemother 56:5907–5915. doi:10.1128/AAC.00770-12
Wolf D, Kalamorz F, Wecke T, Juszczak A, Mäder U, Homuth G, Jordan S, Kirstein J, Hoppert M, Voigt B, Hecker M, Mascher T (2010) In-depth profiling of the LiaR response of Bacillus subtilis. J Bacteriol 192:4680–4693
Zhang XJ, Feng SY, Li ZT, Feng YM (2015) Expression of Helicobacter pylori hspA gene in Lactococcus lactis NICE system and experimental study on its immunoreactivity. Gastroenterol Res Pract 2015:750932. doi:10.1155/2015/750932
Zhou XX, Li WF, Ma GX, Pan YJ (2006) The nisin-controlled gene expression system: construction, application and improvements. Biotechnol Adv 24:285–295. doi:10.1016/j.biotechadv.2005.11.001
Acknowledgments
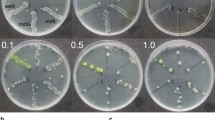
The authors would like to thank Carolin Kobras, Susanne Gebhard, and Anna Staron for providing pictures for Fig. 2. Work in the authors’ laboratory on AMP-responsive 2CSs and their target promoters was financially supported by the Deutsche Forschungsgemeinschaft (to TM).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
This article does not contain any studies with human participants or animals performed by any of the authors.
Funding
Work in the authors’ laboratory on AMP-responsive 2CSs and their target promoters was financially supported by the Deutsche Forschungsgemeinschaft (to TM).
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Wolf, D., Mascher, T. The applied side of antimicrobial peptide-inducible promoters from Firmicutes bacteria: expression systems and whole-cell biosensors. Appl Microbiol Biotechnol 100, 4817–4829 (2016). https://doi.org/10.1007/s00253-016-7519-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-016-7519-3