Abstract
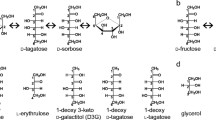
L-Ribose isomerase from Cellulomonas parahominis MB426 (CpL-RI) can catalyze the isomerization between L-ribose and L-ribulose, which are non-abundant in nature and called rare sugars. CpL-RI has a broad substrate specificity and can catalyze the isomerization between D-lyxose and D-xylulose, D-talose and D-tagatose, L-allose and L-psicose, L-gulose and L-sorbose, and D-mannose and D-fructose. To elucidate the molecular basis underlying the substrate recognition mechanism of CpL-RI, the crystal structures of CpL-RI alone and in complexes with L-ribose, L-allose, and L-psicose were determined. The structure of CpL-RI was very similar to that of L-ribose isomerase from Acinetobacter sp. strain DL-28, previously determined by us. CpL-RI had a cupin-type β-barrel structure, and the catalytic site was detected between two large β-sheets with a bound metal ion. The bound substrates coordinated to the metal ion, and Glu113 and Glu204 were shown to act as acid/base catalysts in the catalytic reaction via a cis-enediol intermediate. Glu211 and Arg243 were found to be responsible for the recognition of substrates with various configurations at 4- and 5-positions of sugar. CpL-RI formed a homo-tetramer in crystals, and the catalytic site independently consisted of residues within a subunit, suggesting that the catalytic site acted independently. Crystal structure and site-direct mutagenesis analyses showed that the tetramer structure is essential for the enzyme activity and that each subunit of CpL-RI could be structurally stabilized by intermolecular contacts with other subunits. The results of growth complementation assays suggest that CpL-RI is involved in a novel metabolic pathway using L-ribose as a carbon source.





Similar content being viewed by others
References
Allen KN, Lavie A, Glasfeld A, Tanada TN, Gerrity DP, Carlson SC, Farber GK, Petsko GA, Ringe D (1994) Role of the divalent metal ion in sugar binding, ring opening, and isomerization by D-xylose isomerase: replacement of a catalytic metal by an amino acid. Biochemistry 33:1488–1494. doi:10.1021/bi00172a027
Anderson RL, Allison DP (1965) Purification and characterization of D-lyxose isomerase. J Biol Chem 240:2367–2372
Brunger AT (1993) X-PLOR 3.1: A System for X-Ray Crystallography and NMR. Yale University Press, New Haven
Cheng YC (2001) Potential use of antiviral L-nucleoside analogues for the prevention or treatment of viral associated cancers. Cancer Lett 162:S33–S37. doi:10.1016/S0304-3835(00)00650-9
Cho EA, Lee DW, Cha YH, Lee SJ, Jung HC, Pan JG, Pyun YR (2007) Characterization of a novel D-lyxose isomerase from Cohnella laevoribosii RI-39 sp. J Bacteriol 189:1655–1663. doi:10.1128/JB.01568-06
Choi JG, Hong SH, Kim YS, Kim KR, Oh DK (2012) Characterization of a recombinant thermostable D-lyxose isomerase from Dictyoglomusturgidum that produces D-lyxose from D-xylulose. Biotechnol Lett 34:1079–1085. doi:10.1007/s10529-012-0874-y
Collins P, Ferrier R (1995) Monosaccharides: Their Chemistry and Their Roles in Natural Products. Wiley, West Sussex
Davenport RC, Bash PA, Seaton BA, Karplus M, Petsko GA, Ringe D (1991) Structure of the triosephosphate isomerase-phosphoglycolohydroxamate complex: an analogue of the intermediate on the reaction pathway. Biochemistry 30:5821–5826. doi:10.1021/bi00238a002
DeLano WL (2002) ThePyMOL Molecular Graphics System. DeLano Scientific, San Carlos, CA, USA, http://www.pymol.org
Emsley P, Cowtan K (2004) Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi:10.1107/S0907444904019158
Gumina G, Song GY, Chu CK (2001) L-Nucleosides as chemotherapeutic agents. FEMS Microbiol Lett 202:9–15. doi:10.1111/j.1574-6968.2001.tb10773.x
Gunsalus IC, Horecker BL, Wood WA (1955) Pathways of carbohydrate metabolism in microorganisms. Bacteriol Rev 19:79–128
Hayashi N, Iida T, Yamada T, Okuma K, Takehara I, Yamamoto T, Yamada K, Tokuda M (2010) Study on the postprandial blood glucose suppression effect of D-psicose in borderline diabetes and the safety of long-term ingestion by normal human subjects. Biosci Biotechnol Biochem 74:510–519. doi:10.1271/bbb.90707
Hayashi N, Yamada T, Takamine S, Iida T, Okuma K, Tokuda M (2014) Weight reducing effect and safety evaluation of rare sugar syrup by a randomized double-blind, parallel-group study in human. J Funct Foods 11:152–159. doi:10.1016/j.jff.2014.09.020
Heath EC, Hurwitz J, Horecker BL, Ginsburg A (1958) Pentose fermentation by Lactobacillus plantarum. J Biol Chem 231:1009–1029
Izumori K, Yamanaka K, Elbein AD (1976) Pentose metabolism in Mycobacterium smegmatis: specificity of induction of pentose isomerases. J Bacteriol 128:587–591
Kim K, Kim HJ, Oh DK, Cha SS, Rhee S (2006) Crystal structure of D-psicose 3-epimerase from Agrobacterium tumefaciens and its complex with true substrate D-fructose: a pivotal role of metal in catalysis, an active site for the non-phosphorylated substrate, and its conformational changes. J Mol Biol 361:920–931. doi:10.1016/j.jmb.2006.06.069
Korndörfer IP, Fessner WD, Matthews BW (2000) The structure of rhamnose isomerase from Escherichia coli and its relation with xylose isomerase illustrates a change between inter and intra-subunit complementation during evolution. J Mol Biol 300:917–933. doi:10.1006/jmbi.2000.3896
Krissinel E, Henrick K (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372:774–797. doi:10.1016/j.jmb.2007.05.022
Kwon HJ, Yeom SJ, Park CS, Oh DK (2010) Substrate specificity of a recombinant D-lyxose isomerase from Providenciastuartii for monosaccharides. J Biosci Bioeng 110:26–31. doi:10.1016/j.jbiosc.2009.12.011
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1992) PROCHECK v.2: Programs to Check the Stereochemical Quality of Protein Structures. Oxford Molecular Ltd, Oxford
Lee N, Gielow W, Martin R, Hamilton E, Fowler A (1986) The organization of the araBAD operon of Escherichia coli. Gene 47:231–244. doi:10.1016/0378-1119(86)90067-3
Levin GV (2002) Tagatose, the new GRAS sweetener and health product. J Med Food 5:23–36. doi:10.1089/109662002753723197
Marles-Wright J, Lewis RJ (2011) The structure of a D-lyxoseisomerase from the σBregulon of Bacillus subtilis. Proteins 79:2015–2019. doi:10.1002/prot.23028
McRee DE (1999) XtalView/Xfit: a versatile program for manipulating atomic coordinate and electron density. J Struct Biol 125:156–165. doi:10.1006/jsbi.1999.4094
Mizanur RM, Takata G, Izumori K (2001) Cloning and characterization of a novel gene encoding L-ribose isomerase from Acinetobacter sp. strain DL-28 in Escherichia coli. Biochim Biophys Acta 1521:141–145. doi:10.1016/S0167-4781(01)00290-1
Morimoto K, Terami Y, Maeda Y, Yoshihara A, Takata G, Izumori K (2013) Cloning and characterization of the L-ribose isomerase gene from Cellulomonas parahominis MB426. J Biosci Bioeng 115:377–381. doi:10.1016/j.jbiosc.2012.10.021
Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of Macromolecular Structures by the Maximum-Likelihood Method. Acta Cryst D Biol Crystallogr 53:240–255. doi:10.1107/S0907444996012255
Otwinowski Z, Minor W (1997) Processing of X-ray Diffraction Data Collected in Oscillation Mode. Method in Enzymology 276: Macromolecular Crystallography part A 307–326. doi:10.1016/s0076-6879(97)76066-x
Park CS, Kwon HJ, Yeom SJ, Oh DK (2010) Mannose production from fructose by free and immobilized D-lyxose isomerases from Providenciastuartii. Biotechnol Lett 32:1305–1309. doi:10.1007/s10529-010-0300-2
Patel DH, Wi SG, Lee SG, Lee DS, Song YH, Bae HJ (2011) Substrate specificity of the Bacillus licheniformis lyxose isomerase YdaE and its application in in vitro catalysis for bioproduction of lyxose and glucose by two-step isomerization. Appl Environ Microbiol 77:3343–3350. doi:10.1128/AEM. 02693-10
Ramachandran GN, Sasisekharan V (1968) Conformation of polypeptides and protein. Advan Protein Chem 23:283–437
Schleif R (2000) Regulation of the L-arabinose operon of Escherichia coli. Trends Genet 16:559–565. doi:10.1016/S0168-9525(00)02153-3
Shimonishi T, Izumori K (1986) A new enzyme, L-ribose isomerase from Acinetobacter sp. J Ferment Bioeng 81:493–497. doi:10.1016/0922-338X(96)81468-1
Staalduinen LM, Park CS, Yeom SJ, Adams-Cioaba MA, Oh DK, Jia Z (2010) Structure-based annotation of a novel sugar isomerase from the pathogenic E. coli O157:H7. J Mol Biol 401:866–881. doi:10.1016/j.jmb.2010.06.063
Stevens FJ, Wu TT (1976) Growth on D-lyxose of a mutant strain of Escherichia coli K12 using a novel isomerase and enzymes related to D-xylose metabolism. J Gen Microbiol 97:257–265. doi:10.1099/00221287-97-2-257
Takeda K, Yoshida H, Izumori K, Kamitori S (2010) X-ray structures of Bacillus pallidusD-arabinose isomerase and its complex with L-fucitol. Biochim Biophys Acta 1804:1359–1368. doi:10.1016/j.bbapap.2010.01.018
Vagin A, Teplyakov A (1997) MOLREP: An automated program for molecular replacement. J Appl Cryst 30:1022–1025. doi:10.1107/S0907444909042589
Whitlow M, Howard AJ, Finzel BC, Poulos TL, Winborne E, Gilliland GL (1991) Ametalmediated hydride shift mechanism for xylose isomerase based onthe 1.6 Å Streptomyces rubiginosusstructures with xylitol and D-xylose. Proteins Struct Funct Genet 9:153–173. doi:10.1002/prot.340090302
Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67:235–242. doi:10.1107/S0907444910045749
Yoshida H, Yamada M, Ohyama Y, Takada G, Izumori K, Kamitori S (2007) The structures of L-rhamnose isomerase from Pseudomonas stutzeri in complexes with L-rhamnose and D-allose provide insights into broad substrate-specificity. J Mol Biol 365:1505–1516. doi:10.1016/j.jmb.2006.11.004
Yoshida H, Teraoka M, Yoshihara A, Izumori K, Kamitori S (2011) Overexpression, crystallization and preliminary X-ray diffraction analysis of L-ribose isomerase from Acinetobacter sp. strain DL-28. Acta Crystallogr Sect F Struct Biol Cryst Commun 67:1281–1284. doi:10.1107/S1744309111030351
Yoshida H, Yoshihara A, Teraoka M, Terami Y, Takata G, Izumori K, Kamitori S (2014) X-ray structure of a novel L-ribose isomerase acting on a non-natural sugar L-ribose as its ideal substrate. FEBS J 281:3150–3164. doi:10.1111/febs.12850
Acknowledgments
We thank Dr. A. Itoh and Mr. K. Yube for their technical assistant with DNA sequencing. Y. Terami is supported by a Grant-in-Aid from the Japan Society for the Promotion of Science (JSPS) Research Fellows. This study was supported in part by Grants-in-Aid for Scientific Research (25440028, 23770122, 22780068, 26450097, 22580088) from JSPS, and by the funds from the Kagawa University New Research Areas and Collaborative Research Organization. This research was performed with the approval of the Photon Factory Advisory Committee and National Laboratory for High Energy Physics (2012G550 and 2013G506) Japan.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 1714 kb)
Rights and permissions
About this article
Cite this article
Terami, Y., Yoshida, H., Uechi, K. et al. Essentiality of tetramer formation of Cellulomonas parahominis L-ribose isomerase involved in novel L-ribose metabolic pathway. Appl Microbiol Biotechnol 99, 6303–6313 (2015). https://doi.org/10.1007/s00253-015-6417-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-015-6417-4