Abstract
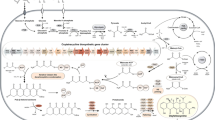
Titer improvement is a constant requirement in the fermentation industry. The traditional method of “random mutation and screening” has been very effective despite the considerable amount of time and resources it demands. Rational metabolic engineering, with the use of recombinant DNA technology, provides a novel, alternative strategy for titer improvement that complements the empirical method used in industry. Manipulation of the specific regulatory systems that govern secondary metabolite production is an important aspect of metabolic engineering that can efficiently improve fermentation titers. In this review, we use examples from Streptomyces secondary metabolism, the most prolific source of clinically used drugs, to demonstrate the power and utility of exploiting natural regulatory networks, in particular pathway-specific regulators, for titer improvement. Efforts to improve the titers of fredericamycin, C-1027, platensimycin, and platencin in our lab are highlighted.


Similar content being viewed by others
References
Adrio JL, Demain AL (2006) Genetic improvement of process yielding microbial products. FEMS Microbiol Rev 30:187–214
Bibb MJ (2005) Regulation of secondary metabolism in streptomycetes. Curr Opin Microbiol 8:208–215
Borodina I, Krabben P, Nielsen J (2005) Genome-scale analysis of Streptomyces coelicolor A3(2) metabolism. Genome Res 15:820–829
Bruheim P, Sletta H, Bibb MJ, White J, Levine DW (2002) High-yield actinorhodin production in fed-batch culture by a Streptomyces lividans strain overexpressing the pathway-specific activator gene actll-ORF4. J Ind Microbiol Biotech 28:103–111
Chen Y, Luo Y, Ju J, Wendt-Pienkowski E, Rajski SR, Shen B (2008a) Identification of fredericamycin E from Streptomyces griseus: insight into fredericamycin A biosynthesis highlighting carbaspirocycle formation. J Nat Prod 71:431–437
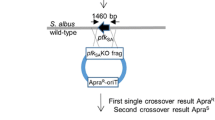
Chen Y, Wendt-Pienkowski E, Shen B (2008b) Identification and utility of FdmR1 as a Streptomyces antibiotic regulatory protein activator for fredericamycin production in Streptomyces griseus ATCC 49344 and heterologous hosts. J Bacteriol 190:5587–5596
Chen Y, Wendt-Pienkowski E, Rajski S, Shen B (2009) In vivo investigation of the roles of FdmM and FdmM1 in fredericamycin biosynthesis unveiling a new family of oxygenases. J Biol Chem 284:24735–24743
Cropp A, Chen S, Liu H, Zhang W, Reynolds KA (2001) Genetic approaches for controlling ratios of related polyketide products in fermentation processes. J Ind Microbiol Biotech 27:368–377
Dangel V, Eustáquio AS, Gust B, Heide L (2008) novE and novG act as positive regulators of novobiocin biosynthesis. Arch Microbiol 190:509–519
Demain AL (2006) From natural products discovery to commercialization: a success story. J Ind Microbiol Biotech 33:486–495
Eustáquio AS, Li SM, Heide L (2005) NovG, a DNA-binding protein acting as a positive regulator of novobiocin biosynthesis. Microbiology 151:1946–1961
Feng Z, Wang L, Rajski SR, Xu Z, Coeffet-LeGal MF, Shen B (2009) Engineered production of iso-migrastatin in heterologous Streptomyces hosts. Bioorg Med Chem 17:2147–2153
Gallegos MT, Schleif R, Bairoch A, Hofmann K, Ramos JL (1997) AraC/Xyls family of transcriptional regulators. Microbiol Mol Biol Rev 61:393–410
Galm U, Shen B (2006) Expression of biosynthetic gene clusters in heterologous hosts for natural product production and combinatorial biosynthesis. Exp Opin Drug Discov 1:409–437
Gust B, Challis GL, Fowler K, Kieser T, Chater KF (2003) PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci U S A 100:1541–1546
Haydon DJ, Guest JR (1991) A new family of bacterial regulatory proteins. FEMS Microbiol Lett 63:291–295
Hillerich B, Westpheling J (2006) A new GntR family transcriptional regulator in Streptomyces coelicolor is required for morphogenesis and antibiotic production and controls transcription of an ABC transporter in response to carbon source. J Bacteriol 188:7477
Hertweck C (2009) The biosynthetic logic of polyketide diversity. Angew Chem Int Ed 48:4688–4716
Horsman GP, Van Lanen SG, Shen B (2009) Iterative type I polyketide synthases for enediyne core biosynthesis. Methods Enzymol 459:97–112
Jung WS, Jeong SJ, Park SR, Choi CY, Park BC, Park JW, Yoon YJ (2008) Enhanced heterologous production of desosaminyl macrolides and their hydroxylated derivatives by overexpression of the pikD regulatory gene in Streptomyces venezuelae. Appl Environ Microbiol 74:1972–1979
Kitani S, Ikeda H, Sakamoto T, Noguchi S, Nihira T (2009) Characterization of a regulatory gene, aveR, for the biosynthesis of avermectin in Streptomyces avermitilis. Appl Microbiol Biotechnol 82:1089–1096
Koffas M, Roberge C, Lee K, Stephanopoulos G (1999) Metabolic engineering. Annu Rev Biomed Eng 1:535–557
Koglin A, Walsh CT (2009) Structural insights into nonribosomal peptide enzymatic assembly lines. Nat Prod Rep 26:987–1000
Li C, Hazzard C, Florova G, Reynolds KA (2009) High titer production of tetracenomycins by heterologous expression of the pathway in a Streptomyces cinnamonensis industrial monensin producer strain. Metab Eng. doi:10.1016/j.ymben.2009.06.004
Lim SK, Ju J, Zazopoulos E, Jiang H, Seo JW, Chen Y, Feng Z, Rajski SR, Farnet CM, Shen B (2009) iso-Migrastatin, migrastatin, and dorrigocin production in Streptomyces platensis NRRL 18993 is governed by a single biosynthetic machinery featuring an acyltransferase-less type I polyketide synthase. J Biol Chem 284:29746–29756
Liu W, Christenson SD, Standage S, Shen B (2002) Biosynthesis of the enediyne antitumor antibiotic C-1027. Science 297:1170–1173
Liu W, Nonaka K, Nie L, Zhang J, Christenson SD, Bae J, Van Lanen SG, Zazopoulos E, Farnet CM, Yang CF, Shen B (2005) The neocarzinostatin biosynthetic gene cluster from Streptomyces carzinostaticus ATCC 15944 involving two iterative type I polyketide synthases. Chem Biol 12:293–302
Maddocks SE, Oyston PC (2008) Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiol 154:3609–3623
Nielsen J (2001) Metabolic engineering. Appl Microbiol Botechnol 55:263–283
Oh SY, Shin JH, Roe JH (2007) Dual role of OhrR as a repressor and an activator in response to organic hydroperoxides in Streptomyces coelicolor. J Bacteriol 189:6284–6292
Olano C, Lombó F, Méndez C, Salas JA (2008) Improving production of bioactive secondary metabolites in actinomycetes by metabolic engineering. Metab Eng 10:281–292
Parekh S, Vinci VA, Strobel RJ (2000) Improvement of microbial strains and fermentation process. Appl Microbiol Biotechnol 54:287–301
Parakar A, Trefzer A, Chakraburtty R, Stassi D (2003) Stretomyces genetics: a genomic perspective. Crit Rev Biotechnol 23:1–27
Ramos JL, Martínez-Bueno M, Molina-Henares AJ, Terán W, Watanabe K, Zhang X, Gallegos MT, Brennan R, Tobes R (2005) The TetR family of transcriptional repressors. Microbiol Mol Biol Rev 69:326–356
Retzlaff L, Distler J (1995) The regulator of streptomycin gene expression, StrR, of Streptomyces griseus is a DNA binding activator protein with multiple recognition sites. Mol Microbiol 18:151–162
Rigali S, Derouaux A, Giannotta F, Dusart J (2002) Subdivision of the helix-turn-helix GntR family of bacterial regulators in the FadR, HutC, MocR, and YtrA subfamilies. J Biol Chem 277:12507–12515
Rigali S, Titgemeyer F, Barends S, Mulder S, Thomae AW, Hopwood DA, Van Wezel GP (2008) Feast or famine: the global regulator DasR links nutrient stress to antibiotic production by Streptomyces. EMBO Rep 9:670–675
Scherlach K, Hertweck C (2009) Triggering cryptic natural product biosynthesis in microorganisms. Org Biomol Chem 7:1753–1760
Smanski MJ, Peterson RM, Rajski SR, Shen B (2009) Engineered Streptomyces platensis strains that overproduce antibiotics platensimycin and platencin. Antimicrob Agents Chemother 53:1299–1304
Shao RG, Zhen YS (2008) Enediyne anticancer antibiotic lidamycin: chemistry, biology and pharmacology. Anticancer Agents Med Chem 8:123–131
Tang L, Grimm A, Zhang YX, Hutchinson CR (1996) Purification and characterization of the DNA-binding protein DnrI, a transcriptional factor of daunorubicin biosynthesis in Streptomyces peucetius. Mol Microbiol 22:801–813
Wang J, Soisson SM, Young K, Shoop W, Kodali S, Galgoci A, Painter R, Parthasarathy G, Tang YS, Cummings R, Ha S, Dorso K, Motyl M, Jayasuriya H, Ondeyka J, Herath K, Zhang C, Hernandez L, Allocco J, Basilio A, Tormo JR, Genilloud O, Vicente F, Pelaez F, Colwell L, Lee SH, Michael B, Felcetto T, Gill C, Silver LL, Hermes JD, Bartizal K, Barrett J, Schmatz D, Becker JW, Cully D, Singh SB (2006) Platensimycin is a selective FabF inhibitor with potent antibiotic properties. Nature 441:358–361
Wang J, Kodali S, Lee SH, Galgoci A, Painter R, Dorso K, Racine F, Motyl M, Hernandez L, Tinney E, Colletti SL, Herath K, Cummings R, Salazar O, González I, Basilio A, Vicente F, Genilloud O, Pelaez F, Jayasuriya H, Young K, Cully DF, Singh SB (2007) Discovery of platencin, a dual FabF and FabH inhibitor with in vivo antibiotic properties. Proc Natl Acad Sci USA 104:7612–7616
Wang L, Hu Y, Zhang Y, Wang S, Cui Z, Bao Y, Jiang W, Hong B (2009) Role of sgcR3 in positive regulation of enediyne antibiotic C-1027 production of Streptomyces globisporus C-1027. BMC Microbiol 22:9–14
Wendt-Pienkowski E, Huang Y, Zhang J, Li B, Jiang H, Kwon H, Hutchinson CR, Shen B (2005) Cloning, sequencing, analysis, and heterologous expression of the fredericamycin biosynthetic gene cluster from Streptomyces griseus. J Am Chem Soc 127:16442–16452
Wenzel SC, Müller R (2005) Recent developments towards the heterologous expression of complex bacterial natural product biosynthetic pathways. Curr Opin Biotechnol 16:594–606
Wietzorrek A, Bibb M (1997) A novel family of proteins that regulates antibiotic production in streptomycetes appears to contain an OmpR-like DNA-binding fold. Mol Microbiol 25:1177–1184
Acknowledgements
Studies on natural product biosynthesis and engineered described from the Shen laboratories were supported in part by the National Institutes of Health (NIH) grants CA78747, CA106150, and CA113297. M.J.S. is supported in part by NIH grant T32 GM008347.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chen, Y., Smanski, M.J. & Shen, B. Improvement of secondary metabolite production in Streptomyces by manipulating pathway regulation. Appl Microbiol Biotechnol 86, 19–25 (2010). https://doi.org/10.1007/s00253-009-2428-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-009-2428-3