Abstract
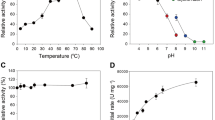
The hemicellulose xylan constitutes a major portion of plant biomass, a renewable feedstock available for conversion to biofuels and other bioproducts. β-xylosidase operates in the deconstruction of the polysaccharide to fermentable sugars. Glycoside hydrolase family 43 is recognized as a source of highly active β-xylosidases, some of which could have practical applications. The biochemical details of four GH43 β-xylosidases (those from Alkaliphilus metalliredigens QYMF, Bacillus pumilus, Bacillus subtilis subsp. subtilis str. 168, and Lactobacillus brevis ATCC 367) are examined here. Sedimentation equilibrium experiments indicate that the quaternary states of three of the enzymes are mixtures of monomers and homodimers (B. pumilus) or mixtures of homodimers and homotetramers (B. subtilis and L. brevis). k cat and k cat/K m values of the four enzymes are higher for xylobiose than for xylotriose, suggesting that the enzyme active sites comprise two subsites, as has been demonstrated by the X-ray structures of other GH43 β-xylosidases. The K i values for d-glucose (83.3–357 mM) and d-xylose (15.6–70.0 mM) of the four enzymes are moderately high. The four enzymes display good temperature (K t 0.5 ∼ 45 °C) and pH stabilities (>4.6 to <10.3). At pH 6.0 and 25 °C, the enzyme from L. brevis ATCC 367 displays the highest reported k cat and k cat/K m on natural substrates xylobiose (407 s−1, 138 s−1 mM−1), xylotriose (235 s−1, 80.8 s−1 mM−1), and xylotetraose (146 s−1, 32.6 s−1 mM−1).




Similar content being viewed by others
References
Brunzelle JS, Jordan DB, McCaslin DR, Olczak A, Wawrzak Z (2008) Structure of the two-subsite β-d-xylosidase from Selenomonas ruminantium in complex with 1,3-bis[tris(hydroxymethyl)methylamino]propane. Arch Biochem Biophys 474:157–166. doi:10.1016/j.abb.2008.03.007
Brüx C, Ben-David A, Shallom-Shezifi D, Leon M, Niefind K, Shoham G, Shoham Y, Schomburg D (2006) The structure of an inverting GH43 β-xylosidase from Geobacillus stearothermophilus with its substrate reveals the role of the three catalytic residues. J Mol Biol 359:97–109. doi:10.1016/j.jmb.2006.03.005
Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B (2009) The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res 37:D233–D238. doi:10.1093/nar/gkn663
Demeler B (2004) UltraScan version 9.9. A comprehensive data analysis software package for analytical ultracentrifugation experiments. The University of Texas Health Science Center, Department of Biochemistry, San Antonio, TX
Dodd D, Cann IK (2009) Enzymatic deconstruction of xylan for biofuel production. Glob Change Biol Bioenergy 1:2–17. doi:10.1111/j.1757-1707.2009.01004.x
Ducret A, Van Oostveen I, Eng JK, Yates JR III, Aebersold R (1998) High throughput protein characterization by automated reverse-phase chromatography/electrospray tandem mass spectrometry. Protein Sci 7:706–719. doi:10.1002/pro.5560070320
Fan Z, Yuan L, Jordan DB, Wagschal K, Heng C, Braker JD (2010) Engineering lower inhibitor affinities in β-d-xylosidase. Appl Microbiol Biotechnol 86:1099–1113. doi:10.1007/s00253-009-2335-7
Himmel ME (ed) (2008) Biomass recalcitrance: deconstructing the plant cell wall for bioenergy. Blackwell Publishing, Oxford
Jordan DB (2008) β-d-Xylosidase from Selenomonas ruminantium: catalyzed reactions with natural and artificial substrates. Appl Biochem Biotechnol 146:137–149. doi:10.1007/s12010-007-8064-4
Jordan DB, Braker JD (2007) Inhibition of the two-subsite β-d-xylosidase from Selenomonas ruminantium by sugars: competitive, noncompetitive, double binding, and slow binding modes. Arch Biochem Biophys 465:231–246. doi:10.1016/j.abb.2007.05.016
Jordan DB, Braker JD (2009) β-d-Xylosidase from Selenomonas ruminantium: thermodynamics of enzyme-catalyzed and noncatalyzed reactions. Appl Biochem Biotechnol 155:330–346. doi:10.1007/s12010-008-8397-7
Jordan DB, Braker JD (2010) β-d-Xylosidase from Selenomonas ruminantium: role of glutamate 186 in catalysis revealed by site-directed mutagenesis, alternate substrates, and active-site inhibitor. Appl Biochem Biotechnol 161:395–410. doi:10.1007/s12010-009-8874-7
Jordan DB, Braker JD (2011) Opposing influences by subsite −1 and subsite +1 residues on relative xylopyranosidase/arabinofuranosidase activities of bifunctional β-d-xylosidase/α-l-arabinofuranosidase. Biochim Biophys Acta 1814:1648–1657. doi:10.1016/j.bbapap.2011.08.010
Jordan DB, Li X-L (2007) Variation in relative substrate specificity of bifunctional β-d-xylosidase/α-l-arabinofuranosidase by single-site mutations: roles of substrate distortion and recognition. Biochim Biophys Acta 1774:1192–1198. doi:10.1016/j.bbapap.2007.06.010
Jordan DB, Wagschal K (2010) Properties and applications of microbial β-d-xylosidases featuring the catalytically efficient enzyme from Selenomonas ruminantium. Appl Microbiol Biotechnol 86:1647–1658. doi:10.1007/s00253-010-2538-y
Jordan D, Li X-L, Dunlap C, Whitehead T, Cotta M (2007) Structure–function relationships of a catalytically efficient β-d-xylosidase. Appl Biochem Biotechnol 141:51–76. doi:10.1007/s12010-007-9210-8
Jordan DB, Mertens JA, Braker JD (2009) Aminoalcohols as probes of the two-subsite active site of β-d-xylosidase from Selenomonas ruminantium. Biochim Biophys Acta 1794:144–158. doi:10.1016/j.bbapap. 2008.09.015
Jordan DB, Wagschal K, Fan Z, Yuan L, Braker JD, Heng C (2011) Engineering lower inhibitor affinities in β-d-xylosidase of Selenomonas ruminantium by site-directed mutagenesis of Trp145. J Ind Microbiol Biotechnol 38:1821–1835. doi:10.1007/s10295-011-0971-2
Jordan DB, Bowman MJ, Braker JD, Dien BS, Hector RE, Lee CC, Mertens JA, Wagschal K (2012) Plant cell walls to ethanol. Biochem J 442:241–252. doi:10.1042/BJ20111922
Leatherbarrow RJ (2001) GraFit version 5. Erithacus Software Limited, Horley, UK
Ly HD, Withers SG (1999) Mutagenesis of glycosidases. Annu Rev Biochem 68:487–522. doi:10.1146/annurev.biochem.68.1.487
Rasmussen LE, Sørensen HR, Vind J, Viksø-Nielsen A (2006) Mode of action and properties of the β-xylosidases from Talaromyces emersonii and Trichoderma reesei. Biotechnol Bioeng 94:869–876. doi:10.1002/bit.20908
Saha BC (2001) Purification and characterization of an extracellular β-xylosidase from a newly isolated Fusarium verticillioides. J Ind Microbiol Biotechnol 27:241–245. doi:10.1038/sj/jim/7000189
Saha BC (2003) Purification and properties of an extracellular β-xylosidase from a newly isolated Fusarium proliferatum. Bioresour Technol 90:33–38. doi:10.1016/S0960-8524(03)00098-1
Speicher KD, Kolbas O, Harper S, Speicher DW (2000) Systematic analysis of peptide recoveries from in-gel digestions for protein identifications in proteome studies. J Biol Tech 11:74–86
Subramaniam S (1998) The Biology Workbench—a seamless database and analysis environment for the biologist. Proteins 32:1–2. doi:10.1002/(SICI)1097-0134(19980701)32:1<1::AID-PROT1>3.0.CO;2-Q
Wagschal K, Franqui-Espiet D, Lee CC, Robertson GH, Wong DW (2005) Enzyme-coupled assay for β-xylosidase hydrolysis of natural substrates. Appl Environ Microbiol 71:5318–5323. doi:10.1128/AEM.71.9.5318-5323.2005
Wagschal K, Franqui-Espiet D, Lee CC, Kibblewhite-Accinelli RE, Robertson GH, Wong DWS (2007) Genetic and biochemical characterization of an α-l-arabinofuranosidase isolated from a compost starter mixture. Enzyme Microb Technol 40:747–753. doi:10.1016/j.enzmictec.2006.06.007
Wagschal K, Heng C, Lee CC, Robertson GH, Orts WJ, Wong DWS (2009a) Purification and characterization of a glycoside hydrolase family 43 β-xylosidase from Geobacillus thermoleovorans IT-08. Appl Biochem Biotechnol 155:304–313. doi:10.1007/s12010-008-8362-5
Wagschal K, Heng C, Lee CC, Wong DWS (2009b) Biochemical characterization of a novel dual-function arabinofuranosidase/xylosidase isolated from a compost starter mixture. Appl Microbiol Biotechnol 81:855–863. doi:10.1007/s00253-008-1662-4
Wagschal K, Jordan DB, Braker JD (2012) Catalytic properties of β-d-xylosidase XylBH43 from Bacillus halodurans C-125 and mutant XylBH43-W147G. Process Biochem 47:366–372. doi:10.1016/j.procbio.2011.07.009
Xu WZ, Shima Y, Negoro S, Urabe I (1991) Sequence and properties of β-xylosidase from Bacillus pumilus IPO. Contradiction of the previous nucleotide sequence. Eur J Biochem 202:1197–1203. doi:10.1111/j.1432-1033.1991.tb16490.x
Yan QJ, Wang L, Jiang ZQ, Yang SQ, Zhu HF, Li LT (2008) A xylose-tolerant β-xylosidase from Paecilomyces thermophila: characterization and its co-action with the endogenous xylanase. Bioresour Technol 99:5402–5410. doi:10.1016/j.biortech.2007.11.033
Yates JR III, Eng JK, McCormack AL, Schieltz D (1995) Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal Chem 67:1426–1436. doi:10.1021/ac00104a020
Yates JR III, Morgan SF, Gatlin CL, Griffin PR, Eng JK (1998) Method to compare collision-induced dissociation spectra of peptides: potential for library searching and subtractive analysis. Anal Chem 70:3557–3565. doi:10.1021/ac980122y
Yoshida S, Hespen CW, Beverly RL, Mackie RI, Cann IKO (2010) Domain analysis of a modular α-l-arabinofuranosidase with a unique carbohydrate binding strategy from the fiber-degrading bacterium Fibrobacter succinogenes S85. J Bacteriol 192:5424–5436. doi:10.1128/JB.00503-10
Acknowledgment
Trypsin digestion and LC-MS/MS analysis was performed at the Wistar Proteomics Facility. Sedimentation equilibrium experiments were conducted at Northwestern University, Keck Biophysics Facility, supported by a Cancer Center Grant (NCI CA060553). Mention of trade names or commercial products in this report is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 713 kb)
Rights and permissions
About this article
Cite this article
Jordan, D.B., Wagschal, K., Grigorescu, A.A. et al. Highly active β-xylosidases of glycoside hydrolase family 43 operating on natural and artificial substrates. Appl Microbiol Biotechnol 97, 4415–4428 (2013). https://doi.org/10.1007/s00253-012-4475-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-012-4475-4