Abstract
Bacillus strains are used for the industrial production of the purine nucleosides inosine and guanosine, which are raw materials for the synthesis of the flavor enhancers disodium inosinate and disodium guanylate. An important precursor of purine nucleosides is 5-phospho-α-d-ribosyl-1-pyrophosphate, which is synthesized by phosphoribosyl pyrophosphate synthetase (PRS, EC 2.7.6.1). Class I PRSs are widespread in bacteria and mammals, are highly conserved among different organisms, and are negatively regulated by two end products of purine biosynthesis, adenosine 5′-diphosphate (ADP) and guanosine 5′-diphosphate (GDP). The D52H, N114S, and L129I mutations in the human PRS isozyme I (PRS1) have been reported to cause uric acid overproduction and gout due to allosteric deregulation and enzyme superactivity. In this study, to find feedback-resistant Bacillus amyloliquefaciens PRS, the influence of the D58H, N120S, and L135I mutations (corresponding to the D52H, N114S, and L129I mutations in PRS1, respectively) on PRS enzymatic properties has been studied. Recombinant histidine-tagged wild-type PRS and three mutant PRSs were expressed in Escherichia coli, purified, and characterized. The N120S and L135I mutations were found to release the enzyme from ADP and GDP inhibition and significantly increase its sensitivity to inorganic phosphate (Pi) activation. In contrast, PRS with the D58H mutation exhibited nearly identical sensitivity to ADP and GDP as the wild-type protein and had a notably greater Pi requirement for activation. The N120S and L135I mutations improved B. amyloliquefaciens and Bacillus subtilis purine nucleoside-producing strains.
Similar content being viewed by others
Introduction
The enzyme ribose-phosphate diphosphokinase or phosphoribosyl pyrophosphate synthetase (PRS; EC 2.7.6.1) catalyzes the biosynthesis of phosphoribosyl pyrophosphate (5-phospho-α-d-ribosyl-1-pyrophosphate, PRPP) from ribose-5-phosphate (R5P) and adenosine 5′-triphosphate (ATP) by transferring the β,γ-diphosphoryl moiety of ATP to the C1-hydroxy group of R5P (Khorana et al. 1958). PRPP is a key metabolic intermediate that is required for the synthesis of purine and pyrimidine nucleotides, the pyridine nucleotide cofactors NAD and NADP, and two amino acids—histidine and tryptophan. In nucleotide synthesis, PRPP is active in the de novo synthesis and in the salvage pathway, which is necessary for the utilization of exogenous purine and pyrimidine bases and nucleosides and their recycling in cells (Fig. 1).
A simplified scheme of the biosynthetic pathways involved in the de novo and salvage synthesis of purine nucleotides in B. subtilis. The black solid lines represent metabolic conversions, the blue dashed lines depict positive regulation of expression, the blue dash-dotted line depicts enzyme activation, and the red dotted lines depict feedback inhibition. Designations: R5P ribose-5-phosphate, PRPP 5-phospho-α-d-ribosyl-1-pyrophosphate, PRA 5-phospho-α-d-ribosylamine, IMP inosine 5′-monophosphate, Hx hypoxanthine, HxR inosine, XMP xanthosine 5′-monophosphate, GMP guanosine 5′-monophosphate, GDP guanosine 5′-diphosphate, GTP guanosine 5′-triphosphate, G guanine, GR guanosine, SAMP succinyladenosine monophosphate, AMP adenosine 5′-monophosphate, ADP adenosine 5′-diphosphate, ATP adenosine 5′-triphosphate, A adenine, AR adenosine
As a central enzyme in the metabolism of nitrogen-containing compounds, PRS is highly conserved among various organisms and is strictly regulated. Three classes of PRSs have been described that differ in dependence on phosphate ions for activity, allosteric regulation mechanisms, and diphosphoryl donor specificity (Arnvig et al. 1990; Fox and Kelley 1971; Gibson et al. 1982; Hove-Jensen et al. 1986; Kadziola et al. 2005; Krath and Hove-Jensen 2001; Switzer and Sogin 1973). The class I PRSs are among the best studied and most widespread in bacteria and mammals. The crystal structures of the Bacillus subtilis PRS (PRSBs) and human PRS isozyme I (PRS1), both of which are the class I PRSs, have been solved (Eriksen et al. 2000, 2002; Li et al. 2007). Despite moderate amino acid sequence identity between PRSBs and PRS1 (47%), the overall structures are similar and possess only a few conformational differences (Li et al. 2007). Class I PRSs require Mg2+ and inorganic phosphate (Pi) for their stability and activity, and the enzymes are sensitive to feedback inhibition by the purine nucleotides adenosine 5′-diphosphate (ADP) and guanosine 5′-diphosphate (GDP) (Arnvig et al. 1990; Gibson et al. 1982; Hove-Jensen et al. 1986; Switzer and Sogin 1973). ADP is the most potent inhibitor, acting in both a competitive and an allosteric manner, with the allosteric inhibition occurring at saturating concentrations of R5P.
Several amino acid substitutions (D52H, N114S, L129I, D183H, A190V, and H193Q) in PRS1 have been described as being responsible for hyperuricemia and gout in humans (Becker et al. 1995; Roessler et al. 1993; Zoref et al. 1975). Genetic and biochemical studies have indicated that the mutant enzymes exhibit increased levels of enzymatic activity due to alterations in the allosteric regulatory mechanisms that result in higher resistance of PRS1 to ADP and GDP inhibition and higher sensitivity to Pi activation. PRS1 superactivity leads to the increased generation of PRPP and, consequently, to acceleration of purine nucleotide biosynthesis that results in accumulation of uric acid, which is the end product of purine metabolism in humans (Zoref et al. 1975).
Sodium salts of inosine 5′-monophosphate (IMP) and guanosine 5′-monophosphate (GMP) are widely used in the food industry as flavor enhancers. IMP and GMP can be produced by Corynebacterium ammoniagenes via direct fermentation (Teshiba and Furuya 1982) or by fermentation to inosine and guanosine followed by subsequent chemical or enzymatic phosphorylation of the nucleosides into nucleotides (Mori et al. 1997). Bacillus strains have been found to have great potential for the industrial production of inosine and guanosine, which are formed in bacterial cells from IMP and GMP, respectively (Fig. 1) (Enei et al. 1976; Ishii and Shiio 1972; Matsui et al. 1982; Miyagawa et al. 1987). PRPP plays a dual positive role in the regulation of the de novo biosynthesis of purine nucleotides in B. subtilis. On the one hand, the PRPP concentration is a limiting factor for the reaction catalyzed by a key regulatory enzyme in this pathway, glutamine PRPP amidotransferase (EC 2.4.2.14). This enzyme, which is encoded by the purF gene, is subject to feedback inhibition by the purine nucleotides, with adenosine 5′-monophosphate (AMP) being the most effective inhibitor. Increasing the PRPP pool has been demonstrated to greatly decrease AMP-mediated glutamine PRPP amidotransferase inhibition (Meyer and Switzer 1979). On the other hand, PRPP binds the PurR repressor and derepresses the PurR-regulated genes that are responsible for the de novo synthesis of purine nucleotides (Weng et al. 1995). The fact that PRPP generation is an essential factor for purine nucleotide biosynthesis and thus for purine nucleoside production indicates that the identification and examination of feedback-resistant PRSs may contribute to the improvement of Bacillus purine nucleoside-producing strains.
In this study, we tested wild-type and mutant Bacillus amyloliquefaciens PRSs to determine which mutations rendered the enzyme insensitive to purine nucleotide inhibition. Wild-type B. amyloliquefaciens PRS and three mutant enzymes containing the D58H, N120S, and L135I mutations (corresponding to the D52H, N114S, and L129I mutations in PRS1, respectively) were expressed in Escherichia coli as histidine-tagged recombinant proteins, purified by immobilized metal affinity chromatography, and biochemically characterized. PRS enzymes with amino acid replacements N120S and L135I exhibited enhanced feedback resistance that resulted in increased purine nucleoside production by B. amyloliquefaciens and B. subtilis producing strains.
Materials and methods
Bacterial strains, bacteriophage, and growth conditions
In this study, several bacterial strains were used: E. coli TG1 [F′ traΔ36 proAB + lacI q lacZΔM15/supE Δ(mcrB-hsdSM)5 thi Δ(lac-proAB)] and BL21(DE3) [F− ompT gal dcm lon hsdS B (r −B m −B ) λ(DE3)], B. subtilis 168 (trpC2) (ATCC 23857) and KMBS375 (ΔdeoD guaB24 Ppur1-Δatt ΔpunA ΔpurA ΔpurR) (Asahara et al. 2010), and B. amyloliquefaciens K wild-type IAM 1523 (Russian National Collection of Industrial Microorganisms (VKPM), B 10986) and AJ1991 (Ade−, Ile−, 8-azaguanineR) (VKPM B 8994). The bacteriophage E40 (VKPM Ph-1629) (Jomantas et al. 1991) was used for plasmid transduction. E. coli and Bacillus cells were grown in Luria–Bertani (LB) or M9 minimal medium (Miller 1972) supplemented with d-glucose (0.4% [w/v] for E. coli or 2% [w/v] for Bacillus). When required, thiamine HCl (5 μg ml−1), amino acids (40 μg ml−1), purine bases (50 μg ml−1), ampicillin (100 μg ml−1), or erythromycin (200 μg ml−1 for E. coli or 5 μg ml−1 for Bacillus) was also added to the medium. Solid medium was generated by adding 18 g l−1 of bacteriological agar to the liquid medium.
Genetic methods
All recombinant DNA manipulations were conducted according to standard procedures (Sambrook and Russell 2001) and the recommendations of the enzyme manufacturer (Fermentas). Plasmid and chromosomal DNA were isolated according to the manufacturer’s instructions using the QIAprep Spin Miniprep Kit (Qiagen) and the GenElute™ Bacterial Genomic DNA Kit (Sigma), respectively.
Transformation of B. subtilis 168 was performed using a previously described standard method (Anagnostopoulos and Spizizen 1961). Introduction of plasmids into the naturally nontransformable AJ1991 was performed by transduction with the E40 bacteriophage prepared from the B. subtilis plasmid-containing intermediate host, as previously described (Zakataeva et al. 2010).
Plasmids were constructed in the E. coli TG1 host. Site-directed mutagenesis was performed using the QuikChange® Site-Directed Mutagenesis kit (Stratagene). Polymerase chain reaction (PCR) amplifications were performed with the Pfu DNA polymerase (Fermentas) for DNA cloning and site-directed mutagenesis and with Taq DNA polymerase (Fermentas) for colony-PCR analysis. All primers used in this study (Supplementary Table 1) were purchased from Syntol (Moscow, Russia). The sequences of all constructs produced by PCR and all of the target chromosome modifications were verified by DNA sequencing using the appropriate primers. Sequence analysis was performed as previously described (Zakataeva et al. 2010).
Cloning of genes encoding wild-type and mutant PRSs from B. amyloliquefaciens and B. subtilis
The thermosensitive shuttle vector pNZT1 (Zakataeva et al. 2010) was used to clone prs from B. amyloliquefaciens and B. subtilis and to incorporate the genetic modifications of these genes into the bacterial chromosomes. The DNA fragment containing the open reading frame (ORF) encoding PRS from B. amyloliquefaciens K (prs Ba) was PCR-amplified using genomic DNA from IAM 1523 as a template and primers (+)gcaD and (−)ctc, which are complementary to the structural portion of the B. subtilis 168 prs-adjacent genes gcaD and ctc, respectively (accession no. NC_000964.3). Sequence analysis of the resulting 2.3-kb PCR product indicated that this DNA fragment contained an ORF (prs Ba) with high nucleotide sequence identity to prs of B. subtilis (85%). Based on the obtained sequence, primers (+)prsSalI and (−)prsPvuII were designed to anneal to the sequence from 511 to 495 bp upstream of the translation start of prs Ba and from 33 to 49 bp downstream of its stop codon, respectively. Using these primers and the genomic DNA from IAM 1523 as a template, prs Ba was PCR-amplified, digested with restriction enzymes SalI and PvuII, and ligated with the SalI–SmaI-digested pNZT1 to produce the pNZT1-PRSBA plasmid.
To clone prs from B. subtilis (prs Bs), a 1.8-kb DNA fragment was PCR-amplified using the primer pair (+)prsBSSalI/(−)prsBS and the genomic DNA from B. subtilis 168 as a template. The amplified DNA was digested with SalI and Ecl136II and cloned into the SalI–SmaI-digested pNZT1 to generate the pNZT1-PRSBS plasmid.
To generate plasmids pNZT1-PRSBA58, pNZT1-PRSBA120, and pNZT1-PRSBA135 containing the D58H (gat to cat), N120S (aac to agc), or L135I (tta to ata) mutations in prs Ba, respectively, site-directed mutagenesis was performed using the plasmid DNA of pNZT1-PRSBA as template and the primer pairs (+)mutBA58/(−)mutBA58, (+)mutBA120/(−)mutBA120, and (+)mutBA135/(−)mutBA135, respectively.
To generate plasmids pNZT1-PRSBS120 and pNZT1-PRSBS135 containing the N120S (aac to agc) or L135I (ctc to ata) mutations in prs Bs, respectively, site-directed mutagenesis was performed using the plasmid DNA of pNZT1-PRSBS as template and the primer pairs (+)mutBS120/(−)mutBS120 and (+)mutBS135/(−)mutBS135, respectively.
Introduction of the mutations into prs of the AJ1991 and KMBS375 chromosomes
The point mutations were introduced into prs of the AJ1991 chromosome using the integrative delivery plasmids pNZT1-PRSBA58, pNZT1-PRSBA120, or pNZT1-PRSBA135 and following a previously described two-step replacement recombination procedure (Zakataeva et al. 2010). Selection of plasmid-free clones with the desired mutation was performed using mismatch-discrimination PCR analysis, as previously described in detail (Zakataeva et al. 2010). To screen prs Ba for the D58H, N120S, or L135I mutations, primer pairs (−)ctctest/(+)testBA58w and (−)ctctest/(+)testBA58m, (+)gcatest/(−)testBA120w and (+)gcatest/(−)testBA120m, or (+)gcatest/(−)testBA135w and (+)gcatest/(−)testBA135m were used, respectively. Introduction of the N120S and L135I mutations into the chromosome of KMBS375 was performed using the delivery plasmids pNZT1-PRSBS120 and pNZT1-PRSBS135 following the same procedure described for AJ1991. The primer pairs (−)prsBS/(+)testBS120w and (−)prsBS/(+)testBS120m and (+)prsBS/(−)testBS135w and (+)prsBS/(−)testBS135m were used to screen prs Bs for the N120S and L135I mutations, respectively. The accuracy of all introduced mutations was confirmed by sequence analysis.
Expression and purification of recombinant PRSBas
To construct the B. amyloliquefaciens PRS expression plasmids, the coding regions of prs Ba and three of its alleles, prs BaD58H, prs BaN120S , and prs BaL135I, which encode enzymes with the D58H, N120S, and L135I mutations, respectively, were PCR-amplified using the primer pair (+)H6-prsNcoI/(−)H6-prsXhoI and the plasmid DNA of pNZT1-PRSBA, pNZT1-PRSBA58, pNZT1-PRSBA120, and pNZT1-PRSBA135 as templates, respectively. The resulting PCR fragments were digested with restriction enzymes NcoI and XhoI and then cloned into the NcoI–XhoI-digested pET15b(+) plasmid (Novagen, Inc.) to create a translation fusion between the polyhistidine-tag sequence of pET15b(+) and the prs Ba, prs BaD58H, prs BaN120S , or prs BaL135I coding region; this procedure yielded the pET15-PRS, pET15-PRS58, pET15-PRS120, or pET15-PRS135 plasmids, respectively.
These plasmids and the vector pET15b(+) were transformed into E. coli BL21(DE3) cells. Cultures of the obtained transformants were grown aerobically in LB medium containing ampicillin at 37 °C until the optical density at 600 nm reached 1.0, and then the cultures were induced with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG; Fermentas) and grown for an additional 3 h at 37 °C. Next, 100 ml of IPTG-induced cells was harvested by centrifugation, washed twice with 0.9% NaCl, suspended in 12 ml of buffer A (20 mM Na2HPO4, 500 mM NaCl, 20 mM imidazole, 2 mM PMSF at pH 7.5), disrupted by sonication, and centrifuged at 10,000 × g at 4 °C for 20 min. The supernatant was applied to a HisTrap HP column (1 ml) (Pharmacia) equilibrated with buffer A. The protein was eluted with scalar concentrations (20 to 500 mM) of imidazole in buffer A using an ÄKTApurifier (GE Healthcare). The PRSBa-containing fractions were combined and applied on a Sephadex G-25 column (Pharmacia) equilibrated with buffer B (50 mM potassium phosphate at pH 7.5, 15% [v/v] glycerol) and eluted in the same buffer. The purified recombinant proteins were aliquoted and stored at −70 °C until required.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), gel filtration, and protein estimation
SDS-PAGE was performed in a 5% stacking gel and a 15% resolving gel, following a previously described method (Laemmli 1970). After electrophoresis, the protein bands were visualized by Coomassie Brilliant Blue R250 staining.
Size-exclusion chromatography was performed with the purified enzyme sample (200 μg) using a Superdex 200 HR 10/30 column (Amersham Biosciences) attached to an ÄKTApurifier (GE Healthcare). The column was equilibrated and eluted with 50 mM potassium phosphate buffer at pH 7.5, containing 5 mM MgCl2 and 0.3 M NaCl at a flow rate of 0.5 ml min−1 at 4 °C. The absorbance of the effluent was monitored at 215, 260, and 280 nm. The column was calibrated with molecular mass standards (Sigma): thyroglobulin (669 kDa), apoferritin (443 kDa), β-amylase (200 kDa), alcohol dehydrogenase (150 kDa), bovine serum albumin (66 kDa), and carbonic anhydrase (29 kDa). The molecular mass of the individual protein was calculated from the elution volumes of the standards (Laurent and Killander 1964). The protein concentrations were determined with the Bradford (1976) protein assay using bovine serum albumin as a standard.
PRS activity assay and kinetic analysis
The purified recombinant PRSBas were diluted in a stabilization buffer containing 50 mM potassium phosphate at pH 7.5, 5 mM MgCl2, 0.03 mM ATP, and 15% (v/v) glycerol. For the evaluation of Pi activation, PRSBas were diluted in a buffer containing 20 mM Tris–HCl at pH 7.5, 10 mM potassium phosphate, 5 mM MgCl2, 0.03 mM ATP, and 15% (v/v) glycerol. Considering the reported properties of the class I PRSs (Arnvig et al. 1990; Eriksen et al. 2000; Hove-Jensen et al. 1986), the activities of the purified recombinant PRSBas were measured in a potassium phosphate buffer in the presence of MgCl2 and saturating concentrations of ATP and R5P. MgCl2 was present at a higher molar concentration than ATP because the enzyme requires both free Mg2+ ions as an activator and the Mg–ATP complex as a substrate (Arnvig et al. 1990). The standard reaction mixture (with a final volume of 0.5 ml) contained 50 mM potassium phosphate buffer at pH 7.5, 1 mM DTT, 1 mM EDTA, 1 mM ATP, 1 mM R5P, 5 mM MgCl2, and 0.03 μg of purified recombinant protein. After preincubation of all components except R5P for 2 min at 37 °C, the reaction was initiated by adding R5P. All reactions were performed at 37 °C for 20 min, as the reaction rate was linear under these conditions. The reaction was terminated by adding of 0.2 M H3PO4 (0.5 ml), followed by centrifugation (10,000 × g, 5 min, 4 °C). A control reaction mixture without R5P was employed for each sample (no AMP accumulation was detected in the control mixture). AMP accumulation was measured by high-performance liquid chromatography (HPLC) using a previously described nonradioactive method (Koshiishi et al. 2001) with modifications. HPLC was performed using an Acquity system supplied with a Photodiode Array Detector 2996 (Waters, USA). The sample was appropriately diluted, injected into an HPLC system equipped with a WAX-1 50 × 4 mm column (Shimadzu, Japan), and eluted at room temperature with 50 mM NaH2PO4 (pH 3.3) at a flow rate of 0.5 ml min−1. The absorbance was measured at 260 nm. The concentration of accumulated AMP was determined using a calibration curve obtained by injecting standard solutions of AMP (Sigma). One unit of activity (U) was defined as 1 μmol of AMP formed per minute under the conditions described above.
To study the kinetic parameters of the purified enzymes, the enzymatic activities were assayed under the conditions described above in standard reaction mixtures, excluding the concentrations of substrates (ATP and R5P) or effectors (ADP, GDP, and Pi). To determine the apparent Michaelis constant (K m) and maximal initial velocity (V max) with respect to ATP and R5P, kinetic analyses were performed with at least seven different concentrations of ATP or R5P in the range of 0.025 to 1 mM, and the concentration of the other substrate was constant at 1 mM. To determine the apparent activation constant (K a), which indicates the Pi concentration leading to half-maximal enzyme activation, the enzyme assay was performed in a standard reaction mixture except that the 50-mM potassium phosphate buffer was replaced with 45 mM Tris–HCl at pH 7.5, and KH2PO4 (Pi) was added at seven different concentrations that varied from 0.5 to 7.0 mM. To determine the IC50 value, which indicates the concentration of the inhibitor needed to inhibit 50% of the protein activity, the enzyme assay was performed in a standard reaction mixture with ADP or GDP added at seven different concentrations that varied from 0 to 1 mM. All measurements were performed in triplicate. To determine the average kinetic parameters, K m, V max, K a, and the Hill coefficient (n H), the measured activities were analyzed using the Lineweaver–Burk plot or the Hill plot with the nonlinear curve-fitting program GraphPad Prism 5 (GraphPad Software Inc., San Diego, CA, USA). The IC50 values were calculated using the measured activities and the GraphPad Prism 5 program.
Determination of growth parameters, glucose consumption, and exogenous purine nucleoside accumulation
Biomass accumulation, glucose consumption, and purine nucleoside production by B. amyloliquefaciens and B. subtilis producing strains were examined using tube fermentations. Cells were cultivated at 34 °C for 18 h in LB medium. Then, 0.3 ml of the obtained seed culture was inoculated into 2.7 ml of a fermentation medium in a 20 × 200-mm test tube and incubated in a rotary shaker at 30 °C for derivatives of AJ1991 and at 34 °C for derivatives of KMBS375 until all of the glucose was exhausted. The fermentation medium for AJ1991 contained the following components (per liter): 60.0 g of glucose, 15.0 g of NH4Cl, 1.0 g of KH2PO4, 0.4 g of MgSO4⋅7H2O, 0.01 g of FeSO4⋅7H2O, 0.01 g of MnSO4⋅4H2O, 0.8 g (as the amount of nitrogen) of soybean meal hydrolysate (Ajinomoto Co, Inc.), 0.3 g of adenine, and 25 g of CaCO3. The fermentation medium for KMBS375 contained the following components (per liter): 30.0 g of glucose, 32.0 g of NH4Cl, 1.0 g of KH2PO4, 0.4 g of MgSO4⋅7H2O, 0.01 g of FeSO4⋅7H2O, 0.01 g of MnSO4⋅4H2O, 0.3 g of dl-methionine, 1.35 g (as the amount of nitrogen) of soybean meal hydrolysate, 0.1 g of adenine, 0.05 g of guanine, and 25 g of CaCO3. Glucose and CaCO3 were sterilized separately. The pH was adjusted to 6.5 before sterilization.
Inosine and guanosine concentrations in the culture broth were determined using HPLC as previously described (Zakataeva et al. 2010). Bacterial growth was assayed by measuring the optical density of the culture broth (OD600) using a spectrophotometer (UV-1800, Shimadzu) at 600 nm. The biomass concentration was calculated from OD600 values using an experimentally determined correlation factor of 0.31 g of cell dry weight (DW) per liter for an OD600 of 1. Glucose concentrations were determined by an enzymatic method using an enzyme electrode (BIOSEN C_line; EKF Diagnostic, Germany). The results represent the mean values with standard deviations from measurements taken from three independent tubes. Glucose-specific and biomass-specific nucleoside yields were defined as the ratio of the total nucleoside produced (grams) to the consumed glucose (grams) or accumulated biomass (grams), respectively. Productivity (grams per liter/hour) was calculated as the concentration of nucleoside produced (gram per liter culture volume) divided by fermentation time.
Statistical analysis
Analysis of variance of all obtained data was performed using the GraphPad Prism 5 program.
Nucleotide sequence accession number
The nucleotide sequence of prs from B. amyloliquefaciens strain K has been deposited in the GenBank nucleotide sequence database under the accession number HQ636460.
Results
Cloning, heterologous expression, and purification of recombinant B. amyloliquefaciens PRSs
The ORF from B. amyloliquefaciens K wild-type strain IAM 1523, which putatively encodes PRS (prs Ba), was cloned, sequenced, and analyzed to illustrate that this gene shared 85% nucleotide sequence identity with prs from B. subtilis. A multiple sequence alignment was generated to compare the deduced amino acid sequence encoded by prs Ba to other class I PRS sequences, and this alignment indicated that PRSBa shares 96% and 47% amino acid identity with PRSBs and PRS1, respectively (Fig. 2). Amino acid residues D52, N114, L129, D183, and A190, identified in PRS1 as being involved in allosteric regulation, are conserved in the B. amyloliquefaciens and B. subtilis PRSs. Thus, the known feedback-resistant mutations in PRS1, aspartic acid to histidine at amino acid residue 52 (D52H), asparagine to serine at amino acid residue 114 (N114S), and leucine to isoleucine at amino acid residue 129 (L129I), are supposed to correspond to the D58H, N120S, and L135I mutations, respectively, in PRSBa and PRSBs.
Amino acid sequence comparison of various class I PRSs. The sequences of the PRSs from B. amyloliquefaciens K strain IAM 1523 (Ba k , accession no. ADU02858.1 [GenBank]) and B. subtilis strain 168 (Bs, accession no. P14193.1 [Swiss-Prot]) and the sequence of human PRS isozyme I (Hum, accession no. CAI42173.1 [GenBank]) are presented. Amino acid conservation is noted as follows: identical residues (black shading, white letters), similar residues (gray shading), and residues altered in feedback-resistant human PRS isozyme I (asterisks). The alignment was performed using the ClustalW program (http://www.ebi.ac.uk/clustalw)
To investigate the basic physical and kinetic properties of the B. amyloliquefaciens PRS and to determine the effect of the D58H, N120S, and L135I mutations on these properties, the recombinant N-terminal polyhistidine-tagged (MGSSHHHHHHSSG) wild-type enzyme (PRSBaWT) and its derivatives with the D58H (PRSBaD58H), N120S (PRSBaN120S), and L135I (PRSBaL135I) mutations were expressed in E. coli BL21(DE3). A protein band with an apparent molecular mass of approximately 35 kDa was detected in supernatants of cells harboring the PRSBa expression plasmids (pET15-PRS, pET15-PRS58, pET15-PRS120, or pET15-PRS135), but not in those harboring empty pET15b(+) vector (for SDS-PAGE, see Supplementary Fig. 1). This value was in agreement with the calculated molecular mass for the recombinant proteins of 36,134 Da (ProtParam, http://cn.expasy.org/tools/protparam.html). Each heterologously produced enzyme was purified from the supernatant of the disrupted cells on nickel-affinity columns to apparent homogeneity as indicated by a single protein band after SDS-PAGE (Supplementary Fig. 1). Three-dimensional structures of PRSBa homologs from B. subtilis and humans, PRSBs and PRS1, respectively (Eriksen et al. 2002; Li et al. 2007; the Protein Data Bank, http://www.pdb.org/), demonstrated that the N-terminal amino acid residues of both enzymes were exposed on the protein surface and were not involved in the formation of catalytic and allosteric sites. Therefore, we predicted that the histidine tag at the N-terminus of PRSBa would not alter the catalytic properties of the enzyme. Indeed, all recombinant histidine-tagged proteins were purified in a soluble, catalytically active form (see below).
The subunit structures of the purified proteins were analyzed by gel filtration. Each of the PRSBaWT, PRSBaD58H, PRSBaN120S, and PRSBaL135I proteins eluted as a single symmetrical peak at a position reflecting molecular masses of 186 ± 20, 184 ± 20, 178 ± 25, and 192 ± 20 kDa, respectively. This result indicated that the enzymes eluted as pentamers (or possibly hexamers).
Biochemical properties of the recombinant PRSBas
The steady-state kinetics of the recombinant PRSBas were studied as a function of R5P and ATP (Mg-ATP). At saturating concentrations of ATP, the kinetic data for all tested enzymes towards R5P fit well to a hyperbolic curve. At saturating concentrations of R5P and varied ATP concentrations, the kinetic data fit to a sigmoidal curve. The apparent V max values of the PRSBaWT and PRSBaD58H proteins were nearly identical (Table 1). The PRSBaN120S and PRSBaL135I proteins exhibited 19–32% higher V max values than those of the PRSBaWT protein under these assay conditions. The apparent K m values for ATP and R5P for PRSBaD58H, PRSBaN120S , and PRSBaL135I were 20–88% higher than those values for the recombinant wild-type enzyme, indicating that the mutant enzymes had lower affinities for ATP and R5P (Table 1).
Similar to class I PRSs from other organisms, it has been demonstrated that B. amyloliquefaciens PRS requires Pi for activity (Fig. 3a). The Pi activation curves of PRSBaWT and three mutant PRSBas were sigmoidal, with K a values of 6.7 and 2.7 mM for PRSBaD58H and PRSBaWT, respectively. K a values of 0.6 mM and less than 0.5 mM were observed for PRSBaN120S and PRSBaL135I, respectively, indicating that the half-activation of PRSBaN120S and PRSBaL135I by Pi occurred at substantially lower concentrations (Table 2). The optimal Pi concentrations for the PRSBaWT, PRSBaD58H, PRSBaN120S, and PRSBaL135I proteins ranged from 30 to 50 mM, and at higher concentrations, an inhibitory effect was observed (data not shown).
Influence of Pi (a), ADP (b), and GDP (c) on the activities of several recombinant PRSBas. The PRS activities of recombinant PRSBaWT (circles), PRSBaD58H (diamonds), PRSBaN120S (triangles), and PRSBaL135I (squares) were measured in the standard reaction mixture, with these exceptions: a 50 mM potassium phosphate buffer at pH 7.5 was replaced with 45 mM Tris–HCl at pH 7.5, and the activator Pi (KH2PO4) was added at the indicated concentrations; b, c inhibitor (ADP or GDP) was added at the indicated concentrations. The enzyme activities were expressed relative to the specific activity of respective enzymes measured in the standard reaction mixture (in the presence of 50 mM Pi and in the absence of ADP and GDP) (Table 2), which were defined as 100%. The values are the means of three independent experiments. The standard deviations were less than 12%
Examination of enzyme inhibition by nucleoside diphosphates indicated that in accordance with the properties of class I PRSs, PRSBaWT was inhibited by GDP and even more efficiently inhibited by ADP (Fig. 3b, c). The mutant enzymes PRSBaN120S and PRSBaL135I proved to be much more resistant to both nucleotides than PRSBaWT, whereas PRSBaD58H exhibited nearly identical sensitivity to ADP and GDP as PRSBaWT (Table 2).
Influence of mutations in PRS of B. amyloliquefaciens and B. subtilis on growth, glucose consumption, and purine nucleoside production
In this study, we were interested in analyzing the effects of the D58H, N120S, and L135I mutations in PRS on the growth and purine nucleoside production by producing strains. A model nucleoside-producing strain AJ1991 was first described as B. subtilis strain G-1136A (Shiro et al. 1965) and later identified as a B. amyloliquefaciens strain (Zakataeva et al. 2005). This strain, which was generated via several rounds of mutagenesis by chemical treatment and UV irradiation, can simultaneously produce inosine and guanosine due to mutations that allow de novo IMP and GMP oversynthesis. There are two primary mutations that give rise to the nucleoside-producing phenotype. The first is a mutation associated with adenine auxotrophy (purA), and the second, yet unidentified, is a mutation that confers resistance to the purine analog 8-azaguanine. The latter mutation caused derepression of IMP synthetic enzymes (Ishii and Shiio 1972). Derivatives of AJ1991, the NZBA58, NZBA120, and NZBA135 strains, containing the D58H, N120S, and L135I mutations in PRS, respectively, were constructed as described in the “Materials and methods” section. Cell growth, glucose consumption, and purine nucleoside production by AJ1991, NZBA58, NZBA120, and NZBA135 were examined during tube fermentation (Fig. 4a, c). The N120S and L135I mutations increased the rate of inosine and guanosine production and total inosine and guanosine accumulation (Fig. 4c), whereas the L135I mutation conferred a more robust phenotype. Compared with AJ1991, glucose-specific inosine and guanosine yields were increased in NZBA120 by 21% and 15%, respectively, whereas those in NZBA135 were increased by 118% and 48%, respectively (Table 3). In addition, the N120S mutation and, to a greater extent, the L135I mutation in AJ1991 caused a small but reliable increase in biomass accumulation, whereas the glucose consumption rates of NZBA120 and NZBA135 were slightly reduced (Fig. 4a). Nevertheless, the biomass-specific inosine yields of NZBA120 and NZBA135 were 19% and 61% higher, respectively, than the yield of the parental strain, and the inosine productivity of NZBA120 and NZBA135 exceeded that of the parental strain by 23% and 81%, respectively (Table 3). By contrast, the D58H mutation in AJ1991 had a negative effect on nucleoside production. Strain NZBA58 demonstrated 12% and 19% reductions in glucose-specific inosine and guanosine yields, respectively, as compared to the parental strain yields.
Influence of various mutations of PRS on growth, glucose consumption, and nucleoside production by B. amyloliquefaciens AJ1991 (a, c) and B. subtilis KMBS375 (b, d). OD600, glucose and nucleoside (inosine and guanosine) concentrations for strains expressing wild-type (circles), mutant D58H (diamonds), mutant N120S (triangles), and mutant L135I PRS (squares) are shown. OD600 and inosine concentrations are shown as filled symbols. Glucose and guanosine concentrations are shown as open symbols. The values are the means of three independent samples. The standard deviations were less than 5% for nucleoside and glucose concentrations and less than 8% for OD600 values
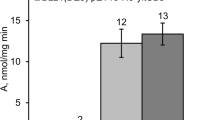
To study the influence of the N120S and L135I mutations in PRS on the nucleoside production of another important industrial bacterium, B. subtilis, the respective mutations were introduced into prs Bs of the KMBS375 chromosome, yielding strains NZBS120 and NZBS135, respectively. Strain KMBS375 (ΔdeoD guaB24 Ppur1-Δatt ΔpunA ΔpurA ΔpurR) was previously constructed via metabolic engineering of B. subtilis strain 168 to achieve inosine production (Asahara et al. 2010). The biomass accumulation, glucose consumption, and nucleoside production of the NZBS120 and NZBS135 strains were compared with those of KMBS375 using the tube fermentation test (Fig. 4b, d). The N120S and L135I mutations in the PRS of KMBS375 caused a slight increase in biomass accumulation and glucose consumption rate. Moreover, as in the case of prs mutants of AJ1991, the purine nucleoside production was increased in both the N120S and L135I mutants of KMBS375. Compared with the parental strain values, the glucose-specific inosine yields in NZBS120 and NZBS135 were 31% and 36% higher, respectively, and the biomass-specific inosine yields were 12% and 19% higher, respectively (Table 3). KMBS375 also accumulated small concentrations of guanosine in the culture broth. Compared with the yields of KMBS375, the glucose-specific guanosine yields for strains NZBS120 and NZBS135 were 61% and 71% higher, respectively.
Discussion
The biochemical properties of several class I PRSs, which catalyze the formation of PRPP from R5P and ATP, have been thoroughly investigated (Arnvig et al. 1990; Becker et al. 1995; Hove-Jensen et al. 1986, 2005; Switzer and Sogin 1973). However, to the best of our knowledge, the B. amyloliquefaciens PRS has not been described. To characterize the PRS of this bacteria, we expressed the recombinant histidine-tagged wild-type PRSBa in E. coli, purified the protein, and studied its biochemical properties. Similar to other class I PRSs, PRSBaWT exhibited sensitivity to ADP and GDP and required Pi for its activity (Fig. 3). The IC50 values for ADP and GDP of PRSBaWT (Table 2) indicated that the B. amyloliquefaciens recombinant enzyme was twofold more sensitive to inhibition by ADP than the PRS from the closely related B. subtilis (Arnvig et al. 1990) and four- and tenfold more resistant to ADP and GDP, respectively, compared to the human PRS isozyme I (Becker et al. 1995).
It has been reported that overexpression of ADP-resistant PRS from E. coli increases inosine accumulation by the E. coli nucleoside-producing strain (Shimaoka et al. 2007). In the engineered Ashbya gossypii riboflavin-producing strain, overexpression of the wild-type and deregulated mutant PRSs significantly enhanced the production of this vitamin, apparently due to excessive synthesis of the riboflavin precursor, GTP (Jiménez et al. 2008). In the B. subtilis riboflavin-producing strain, co-overexpression of wild-type prs and ywlF increased riboflavin accumulation in fed-batch fermentation (Shi et al. 2009). However, Bacillus deregulated PRSs, and their effect on nucleoside production has not been described. We found that the N120S and L135I mutations in PRSBa increased resistance to ADP and GDP inhibition and significantly reduced the Pi requirement for the enzyme’s activity. Interestingly, the recombinant protein with the D58H mutation (PRSBaD58H) had nearly identical sensitivity to ADP and GDP inhibition as the recombinant wild-type enzyme and a notably higher Pi requirement for activation as compared to its human counterpart. The reason for this discrepancy might be related to differences in three-dimensional protein structures of the B. amyloliquefaciens and human enzymes.
PRS resistance to feedback inhibition enhances the synthesis of the purine precursor, PRPP (Zoref et al. 1975), potentially elevating the accumulation of inosine and guanosine, which are the end products of purine metabolism in purine nucleoside-producing strains. Moreover, fermentation with nucleoside-producing strains is usually performed under Pi-limiting conditions to avoid inhibition of IMP/GMP conversion to inosine/guanosine by the respective nucleotidases, thus allowing greater nucleoside accumulation (Teshiba and Furuya 1989). Therefore, mutations in PRS that confer resistance to allosteric inhibition, while simultaneously providing enzymatic activation at lower Pi concentrations, may contribute to the improvement of the respective nucleoside-producing strains. Indeed, examination of N120S and L135I mutants of the B. amyloliquefaciens and B. subtilis model nucleoside-producing strains via tube fermentation revealed an increase in inosine and guanosine production. These strains accumulated higher biomass concentrations than the respective parental strains, probably due to the higher availability of PRPP for the biosynthesis of pyrimidine and pyridine nucleotides and amino acids histidine and tryptophan. Moreover, in the N120S and L135I mutants, both nucleoside and biomass glucose-specific yields were increased compared to the parental strain yields (Table 3), whereas CO2 production was reduced (unpublished observations). These data suggest that intensive R5P outflow in the nucleoside-producing strains, which possess feedback-resistant PRS and deregulated purine nucleotide biosynthesis, can result in extensive carbon flux through NADPH-generating reactions of the oxidative pentose phosphate pathway. The rise in NADPH can depress the tricarboxylic acid cycle (Lim et al. 2002), resulting in the reduction of CO2 generation and more effective glucose utilization in the constructive metabolism (nucleoside and biomass production).
In summary, for the first time, PRS from B. amyloliquefaciens was heterologously expressed, purified, and biochemically characterized. The influence of several mutations on the enzymatic properties of PRS was studied. Two feedback-resistant mutations in PRS, N120S and L135I, were found to be advantageous for purine nucleoside production by B. amyloliquefaciens and B. subtilis model producing strains. These mutations are expected to be helpful for further improvement of industrially important nucleoside-producing strains of Bacillus.
References
Anagnostopoulos C, Spizizen J (1961) Requirements for transformation in Bacillus subtilis. J Bacteriol 81:741–746
Arnvig K, Hove-Jensen B, Switzer RL (1990) Purification and properties of phosphoribosyl-diphosphate synthetase from Bacillus subtilis. Eur J Biochem 192:195–200
Asahara T, Mori Y, Zakataeva NP, Livshits VA, Yoshida K, Matsuno K (2010) Accumulation of gene-targeted Bacillus subtilis mutations that enhance fermentative inosine production. Appl Microbiol Biotechnol 87:2195–2207. doi:10.1007/s00253-010-2646-8
Becker MA, Smith PR, Taylor W, Mustafi R, Switzer RL (1995) The genetic and functional basis of purine nucleotide feedback-resistant phosphoribosylpyrophosphate synthetase superactivity. J Clin Invest 96:2133–2141
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Enei H, Sato K, Anzai Y, Hirose Y (1976) Method of producing guanosine by fermentation. US3969188
Eriksen TA, Kadziola A, Bentsen AK, Harlow KW, Larsen S (2000) Structural basis for the function of Bacillus subtilis phosphoribosyl-pyrophosphate synthetase. Nat Struct Biol 7:303–308
Eriksen TA, Kadziola A, Larsen S (2002) Binding of cations in Bacillus subtilis phosphoribosyldiphosphate synthetase and their role in catalysis. Protein Sci 11:271–279
Fox IH, Kelley WN (1971) Human phosphoribosylpyrophosphate synthetase. Distribution, purification, and properties. J Biol Chem 246:5739–5748
Gibson KJ, Schubert KR, Switzer RL (1982) Binding of the substrates and the allosteric inhibitor adenosine 5′-diphosphate to phosphoribosylpyrophosphate synthetase from Salmonella typhimurium. J Biol Chem 257:2391–2396
Hove-Jensen B, Harlow KW, King CJ, Switzer RL (1986) Phosphoribosylpyrophosphate synthetase of Escherichia coli. Properties of the purified enzyme and primary structure of the prs gene. J Biol Chem 261:6765–6771
Hove-Jensen B, Bentsen AK, Harlow KW (2005) Catalytic residues Lys197 and Arg199 of Bacillus subtilis phosphoribosyl diphosphate synthase. Alanine-scanning mutagenesis of the flexible catalytic loop. FEBS J 272:3631–3639
Ishii K, Shiio I (1972) Improved inosine production and derepression of purine nucleotide biosynthetic enzymes in 8-azaguanine resistant mutants of Bacillus subtilis. Agric Biol Chem 36:1511–1522
Jiménez A, Santos MA, Revuelta JL (2008) Phosphoribosyl pyrophosphate synthetase activity affects growth and riboflavin production in Ashbya gossypii. BMC Biotechnol 8:67. doi:10.1186/1472-6750-8-67
Jomantas JAV, Fiodorova JA, Abalakina EG, Kozlov YI (1991) Genetics of Bacillus amyloliquefaciens. 6th International Conference on Bacilli, Stanford, CA, abstr. no T7
Kadziola A, Jepsen CH, Johansson E, McGuire J, Larsen S, Hove-Jensen B (2005) Novel class III phosphoribosyl diphosphate synthase structure and properties of the tetrameric, phosphate-activated, non-allosterically inhibited enzyme from Methanocaldococcus jannaschii. J Mol Biol 354:815–828
Khorana HG, Fernandes JF, Kornberg A (1958) Pyrophosphorylation of ribose 5-phosphate in the enzymatic synthesis of 5-phosphorylribose 1-pyrophosphate. J Biol Chem 230:941–948
Koshiishi C, Crozier A, Ashihara H (2001) Profiles of purine and pyrimidine nucleotides in fresh and manufactured tea leaves. J Agric Food Chem 49:4378–4382
Krath BN, Hove-Jensen B (2001) Implications of secondary structure prediction and amino acid sequence comparison of class I and class II phosphoribosyl diphosphate synthases on catalysis, regulation, and quaternary structure. Protein Sci 10:2317–2324
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Laurent TC, Killander J (1964) A theory of gel filtration and its experimental verification. J Chromatogr 14:317–330
Li S, Lu Y, Peng B, Ding J (2007) Crystal structure of human phosphoribosylpyrophosphate synthetase 1 reveals a novel allosteric site. Biochem J 401:39–47
Lim SJ, Jung YM, Shin HD, Lee YH (2002) Amplification of the NADPH-related genes zwf and gnd for the oddball biosynthesis of PHB in an E. coli transformant harboring a cloned phbCAB operon. J Biosci Bioeng 93:543–549
Matsui H, Sato K, Enei H, Takinami K (1982) 5′-Nucleotidase activity in improved inosine-producing mutants of Bacillus subtilis. Agric Biol Chem 46:2347–2352
Meyer E, Switzer RL (1979) Regulation of Bacillus subtilis glutamine phosphoribosylpyrophosphate amidotransferase activity by end products. J Biol Chem 254:5397–5402
Miller JH (1972) Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor
Miyagawa K, Doi M, Akiyama S (1987) Method of producing inosine and/or guanosine. US4701413
Mori H, Iida A, Fujio T, Tetsushiba S (1997) A novel process of inosine 5′-monophosphate production using overexpressed guanosine/inosine kinase. Appl Microbiol Biotechnol 48:693–698
Roessler BJ, Nosal JM, Smith PR, Heidler SA, Palella TD, Switzer RL, Becker MA (1993) Human X-linked phosphoribosylpyrophosphate synthetase superactivity is associated with distinct point mutations in the PRPS1 gene. J Biol Chem 268:26476–26481
Sambrook J, Russell D (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Shi S, Chen T, Zhang Z, Chen X, Zhao X (2009) Transcriptome analysis guided metabolic engineering of Bacillus subtilis for riboflavin production. Metab Eng 11:243–252
Shimaoka M, Takenaka Y, Kurahashi O, Kawasaki H, Matsui H (2007) Effect of amplification of desensitized purF and prs on inosine accumulation in Escherichia coli. J Biosci Bioeng 103:255–261
Shiro T, Konishi S, Tamagawa Y, Maruyama T (1965) Method of producing guanosine by fermentation. US3575809
Switzer RL, Sogin DC (1973) Regulation and mechanism of phosphoribosylpyrophosphate synthetase. V. Inhibition by end products and regulation by adenine diphosphate. J Biol Chem 248:1063–1073
Teshiba S, Furuya A (1982) Mechanisms of 5′-inosinic acid accumulation by permeability mutants of Brevibacterium ammoniagenes. I. Genetical improvement of 5′-IMP productivity of a permeability mutant of B. ammoniagenes. Agric Biol Chem 46:2257–2263
Teshiba S, Furuya A (1989) Production of nucleotides and nucleosides by fermentation. Japanese technology reviews, vol. 3. Gordon and Breach Science, New York
Weng M, Nagy PL, Zalkin H (1995) Identification of the Bacillus subtilis pur operon repressor. Proc Natl Acad Sci USA 92:7455–7459
Zakataeva NP, Livshits VA, Gronskiy SV, Kutukova EK, Novikova AE, Kozlov Yu I (2005) Method for producing purine nucleosides and nucleotides by fermentation using bacterium belonging to the genus Bacillus or Escherichia. WO2005095627
Zakataeva NP, Nikitina OV, Gronskiy SV, Romanenkov DV, Livshits VA (2010) A simple method to introduce marker-free genetic modifications into the chromosome of naturally nontransformable Bacillus amyloliquefaciens strains. Appl Microbiol Biotechnol 85:1201–1209. doi:10.1007/s00253-009-2276-1
Zoref E, De Vries A, Sperling O (1975) Mutant feedback-resistant phosphoribosylpyrophosphate synthetase associated with purine overproduction and gout. Phosphoribosylpyrophosphate and purine metabolism in cultured fibroblasts. J Clin Invest 56:1093–1099
Acknowledgments
We are grateful to R. S. Shakulov for valuable advices and critically reading the manuscript. We thank S. V. Smirnov and S. V. Gronskiy for their help and fruitful discussions.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 10.1 kb)
Rights and permissions
About this article
Cite this article
Zakataeva, N.P., Romanenkov, D.V., Skripnikova, V.S. et al. Wild-type and feedback-resistant phosphoribosyl pyrophosphate synthetases from Bacillus amyloliquefaciens: purification, characterization, and application to increase purine nucleoside production. Appl Microbiol Biotechnol 93, 2023–2033 (2012). https://doi.org/10.1007/s00253-011-3687-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-011-3687-3