Abstract
Background
Phosphoribosyl pyrophosphate (PRPP) is a central compound for cellular metabolism and may be considered as a link between carbon and nitrogen metabolism. PRPP is directly involved in the de novo and salvage biosynthesis of GTP, which is the immediate precursor of riboflavin. The industrial production of this vitamin using the fungus Ashbya gossypii is an important biotechnological process that is strongly influenced by substrate availability.
Results
Here we describe the characterization and manipulation of two genes of A. gossypii encoding PRPP synthetase (AGR371C and AGL080C). We show that the AGR371C and AGL080C gene products participate in PRPP synthesis and exhibit inhibition by ADP. We also observed a major contribution of AGL080C to total PRPP synthetase activity, which was confirmed by an evident growth defect of the Δagl080c strain. Moreover, we report the overexpression of wild-type and mutant deregulated isoforms of Agr371cp and Agl080cp that significantly enhanced the production of riboflavin in the engineered A. gossypii strains.
Conclusion
It is shown that alterations in PRPP synthetase activity have pleiotropic effects on the fungal growth pattern and that an increase in PRPP synthetase enzymatic activity can be used to enhance riboflavin production in A. gossypii.
Similar content being viewed by others
Background
Ashbya gossypii is a filamentous hemiascomycete, which has been considered a paradigm of sustainable "white" biotechnology through its use in the industrial overproduction of riboflavin and other vitamins [1]. During the late growth phase, when maximum biomass has been reached, A. gossypii naturally exhibits high levels of riboflavin production as a detoxifying and protective mechanism [2]. However, we have recently described that riboflavin production can be enhanced considerably by genetic and metabolic engineering of the purine pathway, which provides the precursor for riboflavin biosynthesis [3, 4].
Riboflavin is synthesized from GTP and ribulose 5-phosphate through a multi-step pathway controlled by the RIB genes (RIB1 to RIB5 and RIB7 in A. gossypii) [5]. GTP, the immediate precursor for riboflavin production, is synthesized through the de novo purine pathway (Figure 1), which starts with the formation of PRPP. Alternatively, GTP can be formed through the purine salvage pathways (Figure 1), which recycle purines with the consumption of PRPP [6].
Metabolic contribution of PRPP to the purine biosynthesis and other anabolic pathways. The purine pathway is shaded; the de novo purine pathway starts with PRPP biosynthesis, while salvage pathways use PRPP to transform purine bases to monophosphate nucleotides. Dashed arrows indicate a multi-step pathway; sAMP, adenylosuccinate.
PRPP, synthesized from ribose-5-phosphate and ATP, is therefore a key compound for purine and riboflavin biosynthesis, and it is also an important cellular metabolite because it represents a link between carbon and nitrogen metabolism. PRPP is a biosynthetic precursor of histidine and tryptophan, and it is also required for the de novo and salvage pathways of purine, pyrimidine and pyridine (NAD+, NADP+) nucleotides. It has been calculated that approximately 80% of the metabolic flux through PRPP is directed to purine and pyrimidine synthesis [7].
The formation of PRPP is catalyzed by the enzyme PRPP synthetase, which is encoded by PRS genes. It has been shown that mutations in human PRS genes can either activate or inactivate the enzyme, leading to different hereditary disorders including hyperucemia, mental retardation, developmental delay, and other neurological pathologies [8–11].
According to the Ashbya Genome Database (AGD, http://agd.vital-it.ch/index.html) [12] there are four putative annotated genes encoding PRPP synthetase in A. gossypii. In constrast, Saccharomyeces cerevisiae is equipped with a set of five unlinked PRS genes (PRS1-PRS5) [13, 14]. Thus, in S. cerevisiae PRPP synthetase is organized in two interacting complexes or functional entities: a heterodimer comprising Prs1p-Prs3p; and a heterotrimer consisting of Prs2p-Prs4p-Prs5p [15]. Furthermore, systematic analyses of all possible combinations of PRS deletions in S. cerevisiae have revealed three possible phenotypes: synthetic lethality, when PRS1 or PRS3 deletions are combined with a disruption in PRS5, and the simultaneous deletion of PRS2 and PRS4 in either the Δprs1 or Δprs3 strains. A second phenotype is characterized by growth impairment and reduced enzymatic activity; this phenotype is found in mutants containing disruptions in PRS1 and PRS3, either together or in combination with Δprs2 or Δprs4. Finally a third phenotype, which only consists of a reduction in enzymatic activity, is encountered in single or combined deletions of PRS2, PRS4 and PRS5. This complex scenario can be explained in terms of the existence of three minimal subunits capable of sustaining the required PRPP intracellular pool in S. cerevisiae: namely, Prs1-Prs3; Prs2-Prs5 and Prs4-Prs5 [15].
Based on their enzymatic properties, so far three classes of PRPP synthetases have been described: Class I PRPP synthetases, which are dependent on phosphate ions for activity, are inhibited by purine ribonucleotide diphosphates and exclusively use either ATP or dATP as a diphosphoryl donors. PRPP synthetases from Escherichia coli, S. cerevisiae and mammals belong to class I [16–18]. Class II enzymes are specific from plants and are characterized by their independence of phosphate ions, their lack of allosteric inhibition, and their broad specificity for diphosphoryl donors [19]. Finally, a class III enzyme has recently been described in the archaeon Methanocaldococcus jannaschii, which is activated by phosphate and uses ATP as diphosphoryl donor, but lacks an allosteric site for ADP [20]. In a recent publication, a novel allosteric site for SO42-, whose residues are strictly conserved in eukaryotic class I enzymes has been reported to stabilize the active site of PRPP synthetase [21].
Here we report the characterization of AGR371C and AGL080C genes encoding PRPP synthetases from A. gossypii and their effects on riboflavin production and growth. We found that PRPP synthetases from A. gossypii are inhibited by ADP. Furthermore, by combining AgPRS gene deletions we were able to define three different phenotypes that mimicked those previously encountered in yeast Δprs mutants. Finally, we describe the metabolic engineering of A. gossypii strains whose PRPP synthetase regulatory properties have been modified to significantly enhance the productivity of riboflavin.
Methods
Strains, media and techniques for A. gossypii culture
The A. gossypii ATCC 10895 strain was used and was considered as a wild-type strain. A. gossypii was cultured at 28°C using MA2 rich medium [22], synthetic complete media [23] or synthetic minimal media [24]. A concentration of 250 μg/ml of geneticin (G418) (Sigma, Steinheim, Germany), or 200 μg/ml of hygromycin B (Phytotechnology Laboratories, Shawnee Mission, USA), was used when specified. A. gossypii transformation, genomic DNA and RNA isolation, Southern-blot and northern-blot analyses, spores isolation, cell protein extraction, and HPLC determination of total riboflavin contents were carried out as previously described [24, 25].
PCR-based cloning of AGR371C and AGL080C genes from A. gossypii
Based on the annotated sequences for AGR371C and AGL080C in the AGD database http://agd.vital-it.ch/index.html[12], two pairs of primers were designed to PCR-amplify two genomic regions (approx. 3 Kb each) containing AGR371C- and AGL080C- coding DNA sequences. Both genomic fragments were cloned into a pBluescript-SK+ vector (Stratagene) as KpnI-HindIII (AGR371C) and EcoRI (AGL080C) fragments respectively. The resulting clones were shown to be correct by DNA sequencing of the entire fragments (data not shown).
AGR371C and AGL080C gene disruption and overexpression
For AGR371C and AGL080C disruption, an integration cassette was constructed for each ORF (see below for further details). Briefly, for AGR371C disruption we obtained a kanMX4 selection module for geneticin resistance (G418r) with SalI ends from the plasmid pAG-110 [26], which were subsequently treated with Klenow enzyme (Roche) to generate blunt ends. The kanMX4 blunt-ended fragment was inserted between HincII and EcoRV sites in the AGR371C ORF. The complete replacement module was obtained by digestion with NcoI and KpnI and was used to transform spores of the A. gossypii ATCC 10895 strain. For AGL080C disruption, a hygromycin resistance (Hygr) marker was obtained with BamHI-KpnI ends, which were subsequently treated with Klenow enzyme (Roche). The resulting Hygrmarker was inserted between two EcoRV sites present in the AGL080C ORF. Finally, the AGL080C disruption module was obtained by EcoRI digestion and was used to transform either spores of the A. gossypii ATCC 10895 strain, for the single disruption, or spores of the A. gossypii Δagr371c strain, for the double disruption. Correct integrations were verified by analytical PCR and Southern-blotting experiments.
For the overexpression of different alleles of AGR371C and AGL080C, each ORF was inserted as an NdeI-BamHI fragment into the overexpression cassette described below. Briefly, the overexpression cassette comprised: (i) a selection module for geneticin resistance, (ii) an integration module for stable integration of the cassette into the AgLEU2 locus, and finally (iii) an overexpression module based on the AgGPD promoter and terminator sequences, which have been reported to provide constitutive and high expression levels [3]. The overexpression modules were used to transform spores of the A. gossypii ATCC 10895 strain and positive clones were selected in media containing geneticin. Additionally, positive clones were verified for their leucine auxotrophy and analyzed by Southern-blotting.
PRPP synthetase activity assay
A method previously described by Jensen et al. [27] was used for the quantification of PRPP synthetase activity. We measured the conversion of 32P-labelled ATP into 32P-labelled PRPP according to the following reaction:
32P-ATP + ribose 5-P → 32P-PRPP + AMP
The reaction mix contained 50 mM potassium phosphate, pH 7.5, 1 mM [γ-32P] ATP (10 Ci/mmol), 50 mM triethanolamine, 5 mM ribose 5-P, 5 mM MgCl2, 20 mM NaF, 15 mM phosphoenolpyruvate and 1 μmol/min of pyruvate kinase. We used 5–10 mg of total protein extract and the reaction was incubated at 28°C, taking 10 μl aliquots every 5 min over 1 hour. Samples were mixed with 5 μl of 0.33 M formic acid and immediately applied to a polyethylenimine-cellulose-coated plate (Sigma-Aldrich) for thin-layer chromatography. Separation of radiolabeled ATP and PRPP was carried out in 0.85 M potassium phosphate, pH 3.4. The radiolabeled spots corresponding to ATP and PRPP were cut and the associated radioactivity was quantified by liquid scintillation counting methods.
Site-directed mutagenesis of AGR371C and AGL080C and inhibition of PRPP synthetase
Residue substitutions in the AGR371C and AGL080C ORFs were introduced by site-directed mutagenesis through PCR techniques using the primers listed in the Additional file 1. The inhibition assays were performed with different concentrations of ADP in the PRPP synthetase activity analyses. The inhibition rate was calculated as the percentage of specific activity in the presence of ADP with reference to the absence of ADP.
Results
Cloning and sequence analysis of the AGR371C and AGL080C genes from A. gossypii
We have previously shown that substrate availability is a limiting factor for riboflavin overproduction in A. gossypii and that increases in metabolic flux through the purine pathway significantly enhance riboflavin production [3, 4, 28]. PRPP is an important metabolite for purine biosynthesis because it is required in both the de novo and salvage pathways. This prompted us to wonder whether alterations in the PRPP intracellular pool might affect riboflavin production in A. gossypii.
In the AGD database there are four annotated genes encoding PRPP synthetase in A. gossypii that are syntenic homologues of the PRS genes from S. cerevisiae. A protein sequence alignment of A. gossypii and S. cerevisiae PRPP synthetases revealed a high degree of similarity (see Additional file 2). Furthermore, AGR371C is an ortholog of ScPRS2 and ScPRS4, suggesting that AGR371C is a common ancestor of both ScPRS2 and ScPRS4, which probably originated in a gene duplication event (Fig. 2). Thus, in accordance with the PRPP synthetase interacting complexes described in S. cerevisiae [15], we assumed that two homologous heterodimers might exist in A. gossypii, formed by Aer083cp-Agl080cp and Agr371cp-Adr314cp.
Relationship between the PRS genes from A. gossypii and S. cerevisiae. Phylogenetic tree of the PRS proteins from A. gossypii and S. cerevisiae. The alignment of the protein sequences was performed using the W-CLUSTAL program included in the DNAStar package. In bold AGR371C (AgPRS2,4) and AGL080C (AgPRS3).
In order to study the specific contribution of each heterodimer to the overall PRPP synthetase activity and also to the production of riboflavin, for our next experiments we chose one member of each functional entity (AGR371C and AGL080C). For simplicity, AGR371C and AGL080C are henceforth referred to as PRS2,4 and PRS3, respectively, due to their homology with the S. cerevisiae PRS genes.
We amplified by PCR and cloned two genomic regions where PRS2,4 and PRS3 map and sequenced them. In good agreement with the nucleotide sequence deposited in the AGD database [12], PRS2,4 ORF comprised 957 bp encoding a protein of 318 amino acids, and PRS3 ORF consisted of 963 bp and coded for a protein of 320 amino acids. Both the Prs2,4 and Prs3 proteins contained most of the residues identified as being essential for catalysis, the maintenance of three-dimensional structure or protein-protein interactions in previously characterized PRPP synthetases [21, 29] (see Additional file 2 for details).
Analysis of the 5'-non coding regions of AgPRS2,4 and AgPRS3 using the MatInspector program included in the GenomatixSuite 3.1.1 software http://www.genomatix.de/ unveiled several TATA boxes (not shown) and, more importantly, one putative binding site for the transcription activator Bas1p in both the AgPRS2,4 and AgPRS3 sequences (positions -38 and -197 from ATG, respectively) (not shown). However, our previous work demonstrated that the expression of AgPRS2,4 and AgPRS3 is unaffected upon AgBAS1 deletion, suggesting that the presence of only one binding site is not sufficient to ensure transcriptional regulation by Bas1p [4].
AgPRS2,4 and AgPRS3 gene disruptions affects PRPP synthetase activity and growth
In order to elucidate the precise contribution of AgPRS2,4 and AgPRS3 to the overall PRPP synthetase activity in A. gossypii, we next disrupted both genes by inserting a dominant marker into each ORF. We used a kanMX4 selection module for geneticin resistance (G418r) to disrupt AgPRS2,4 and an hygromycin resistance (Hygr) marker to disrupt AgPRS3 (Fig. 3). The final disruption modules comprised the G418ror Hygrselection cassettes flanked by the AgPRS2,4 or AgPRS3 sequences, respectively, to enable homologous recombination and genomic insertion [30] (Fig. 3). With the disruption modules A. gossypii ATCC 10895 spores were transformed by electroporation as previously described [25]. Homokaryotic G418rand Hygrtransformants were obtained after sporulation and clonal selection of the primary heterokaryotic transformants. AgPRS2,4 and AgPRS3 disruption was confirmed by Southern-blot analysis using specific radiolabeled probes (Fig. 3).
Gene disruption of AgPRS2,4 and AgPRS3. A) Left, schematic representation of the strategy followed to achieve the disruption of the AgPRS2,4 gene (AGR371C). Right, Southern blot analysis to confirm correct PRS2,4 disruption. Genomic DNA was digested with HindIII and a genomic KpnI-HindIII fragment was used as a radioactive probe: lane 1, wild-type strain; lane 2, heterokaryotic disruptant; lane 3, mutant Δprs2,4. B) Left, schematic representation of the strategy for AgPRS3 (AGL080C) gene disruption. Right, Southern blot analysis to verify correct PRS3 disruption. Genomic DNA was digested with SacI and a genomic EcoRI fragment was used as a radioactive probe: lane 1, wild-type strain; lane 2, mutant Δprs3. HIII, HindIII; N, NcoI; HII, HincII; EV, EcoRV; K, KpnI; EI, EcoRI; S, SacI.
None of the Δprs mutants showed any nutritional requirement, confirming that the intracellular PRPP pool in both mutants was sufficient to support growth. Nevertheless, while the mutant agΔprs2,4 did not show any visible phenotype when grown on solid media, the agΔprs3 strain revealed a clear growth alteration, exhibiting mycelium-condensed and smaller colonies (Fig. 4). In addition, agΔprs3 displayed a significant reduction in the sporulation ability, this being 10-fold lower than that of the wild-type strain. We therefore analyzed liquid cultures of both agΔprs2,4 and agΔprs3 and determined the mycelial mass produced along growth. After four days of culture, the agΔprs3 strain displayed an initial growth delay with respect to the wild-type, although after 24 hours of culture its biomass was higher than that of the wild-type strain (Fig. 4).
Growth pattern of Δ prs2,4 and Δ prs3 mutant strains. Left, A. gossypii wild-type and prs mutant strains were grown in liquid MA2 rich medium. At the indicated time-points, mycelia were harvested and weighed. Data are represented as an average of mycelium dry-weight per volume of culture. Error bars represent SD. Right, colony photographs of the wild-type, Δprs2,4 and Δprs3 strains grown on solid MA2 rich medium during 48 hours.
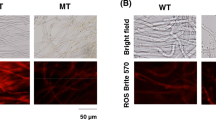
Next we wished to examine the growth alteration of agΔprs3 strain in greater detail by microscopic analysis. Again, agΔprs2,4 and wild-type strains were indistinguishable from each other, both of them showing an initial monopolar hyphal growth and a later bipolar growth, with branched hyphae (Fig. 5). Conversely, Δprs3 disruption caused a shortening on the hyphal length, and an abundant early branching was observed (Fig. 5). This higher branching index could explain the condensed mycelia of the agΔprs3 strain observed on solid media.
PRPP synthetase activity was altered in both agΔprs2,4 and agΔprs3 strains, but disruption of PRS3 resulted in a more marked decrease in the overall enzymatic activity, which in the agΔprs3 strain represented only 14% of the PRPP synthetase activity observed in the wild-type strain. Furthermore, riboflavin production was also partially impaired in the agΔprs3 mutant (Table 1). Taken together, these results suggest a major contribution of the PRS3 paralog to PRPP synthetase activity.
In order to obtain a Δprs2,4-Δprs3 double mutant, spores of the Δprs2,4 strain were transformed with the Δprs3 disruption cassette described above. Selection of primary heterokaryotic clones was made in rich medium containing both geneticin and hygromycin. However, after clonal analysis of 200 homokaryotic strains derived from the previous heterokaryotic clones we failed to find any mutant strain showing resistance to both geneticin and hygromycin, suggesting that the double disruption of PRS2,4 and PRS3 may result in a synthetic lethal phenotype. To verify this hypothesis, we analyzed the initial heterokaryotic transformants (G418r and Hygr) by Southern-blotting and confirmed the presence of both single disruptant Δprs2,4 nuclei and double mutant Δprs2,4-Δprs3 nuclei (not shown). However, none of the homokaryotic transformants analyzed showed the double Δprs2,4-Δprs3 disruption. Additionally, the spores of the heterokaryotic Δprs2,4-Δprs3/Δprs2,4 were isolated by micromanipulation and analyzed individually; the same results described above were obtained. Apparently, the PRPP synthetase activity provided by Δprs2,4 nuclei is sufficient to maintain an adequate pool of PRPP and to allow the survival of the heterokaryotic transformants.
AgPRS2,4 and AgPRS3 overexpression enhances riboflavin production
In a recent paper we reported that PRS2,4 and PRS3 mRNA levels did not change during the trophic and productive phases of A. gossypii growth. Furthermore, PRS2,4 and PRS3 show constitutive low expression levels, suggesting a housekeeping function of these genes [4].
We therefore decided to strongly enhance the expression of PRS2,4 and PRS3 and analyze the effect of their overexpression on riboflavin production. Accordingly, we designed a stable overexpression cassette based on the promoter and terminator sequences of AgGPD (see Materials and Methods for details) (Fig. 6). This cassette allows stable genomic integration into the AgLEU2 locus and selection for G418r. PRS2,4 and PRS3 ORFs were PCR-amplified as NdeI-BamHI fragments and were inserted into the overexpression module. Following this, the final constructs were used to transform spores of the A. gossypii ATCC 10895 strain and heterokaryotic transformants were selected in medium containing geneticin. After sporulation and clonal selection, homokaryotic G418rtransformants were obtained and checked for their leucine auxotrophy. Additionally, integration into the AgLEU2 locus of each overexpression cassette was confirmed by Southern-blotting (Fig. 6).
Overexpression of A. gossypii PRS2,4 and PRS3 genes. Top-left, scheme of the modular cassette used for PRS2,4 and PRS3 overexpression. The integration, selection and overexpression modules are indicated. Top-right, Southern blot analysis to confirm correct integration of the overexpression cassettes into AgLEU2 locus. Genomic DNA of the wild-type (lane 1), GPD-PRS2,4 (lane 2) and GPD-PRS3 (lane 3) strains was digested with PstI and a genomic PstI fragment was used as a radioactive probe. Pm, PmeI; S, SalI; N, NdeI; B, BamHI; Ps, PstI. Bottom, northern blot analysis of A. gossypii total RNA (25 μg) obtained from cultures of the wild-type, GPD-PRS2,4 and GPD-PRS3 strains grown in MA2 rich medium. rRNA 28S was used as a loading control.
Transcriptional analysis of the GPD-PRS2,4 and GPD-PRS3 strains by northern-blotting revealed that mRNA levels of both genes were increased by 30-fold (Fig. 6). However, the increase in PRPP synthetase activity did not correlate with the transcriptional levels (see Table 1), suggesting a regulatory mechanism of the enzymatic activity, as described for other PRPP synthetases [16–18]. Nevertheless, when we quantified the riboflavin yield of the strains overexpressing PRS2,4 and PRS3 we observed a significant improvement in the riboflavin production of both strains (Table 1).
Effect of enzymatic deregulation of AgPRS2,4 and AgPRS3
PRPP synthetases are subject to feed-back regulation by ADP. Furthermore, the amino acids directly involved in inhibitor binding have been elucidated in several PRPP synthetases [21, 29] and it has been reported that specific subtitutions of the leucine 128 and histidine 192 residues of human PRS1 induce allosteric deregulation and enzyme superactivity [9]. In the A. gossypii Prs2,4 and Prs3 proteins, those residues are strictly conserved and correspond to leucines 133 and 132; and histidines 196 and 195 in the Prs2,4 and Prs3 proteins respectively.
We therefore designed mutagenic primers (see Additional file 1) to introduce two point mutations in both PRS2,4 and PRS3 genes. The amino acid substitutions carried out were as follows: the leucines at positions 133 and 132 in Prs2,4 and Prs3, respectively, were substituted by an isoleucine; and the histidines at positions 196 and 195 in Prs2,4 and Prs3 proteins were changed to glutamine. As previously mentioned, both transversions have been reported to cause PRPP synthetase superactivity in humans [9]. After we had amplified the mutant prs2,4 and prs3 ORFs by mutagenic PCR, we cloned the mutant alleles (prs2,4-IQ and prs3-IQ) in the overexpression cassette described above and followed the same strategy to integrate them into the AgLEU2 locus. Correct genomic integrations in homokaryotic transformants were verified by Southern-blotting (not shown).
The functionality of the amino acid substitutions were analyzed by PRPP synthetase enzymatic assays in the prs2,4-IQ and prs3-IQ mutant strains, testing the inhibitory effect of different concentrations of ADP. As shown in figure 6, PRPP synthetase activity was strongly inhibited by ADP in the GPD-PRS2,4 and GPD-PRS3 strains, while the PRPP synthetase activity of the strains carrying the mutant isoforms prs2,4-IQ and prs3-IQ was highly refractory to the inhibitory effect of ADP. Finally, we analyzed the production of riboflavin in the prs2,4-IQ and prs3-IQ strains to compare it with that obtained in the wild-type and also in the strains overexpressing the wild-type PRS2,4 and PRS3 genes. Both strains harboring the deregulated prs2,4-IQ and prs3-IQ attained fairly high production levels, with an 80% greater riboflavin yield than the wild-type strain (Fig. 7). However, riboflavin production did not increase substantially in the prs2,4-IQ and prs3-IQ strains with respect to the yield obtained after the overexpression of the wild-type alleles, suggesting that other regulatory mechanisms may affect the PRPP intracellular pool and riboflavin production.
Effect of overexpression of A. gossypii PRS alleles on PRPP synthetase activity and ADP inhibition. The inhibitory effect of increasing concentrations of ADP was determined using protein extracts from different strains of A. gossypii: GPD-PRS2,4, GPD-PRS3, prs2,4-IQ and prs3-IQ. Results are means of three independent experiments.
Discussion
Riboflavin is currently employed as an additive in human foodstuffs and animal feeds. Indeed, the industrial production of riboflavin exceeds 3000 tons/year, and the chemical production has now been replaced by biotechnological processes using the natural overproducer A. gossypii [28].
Riboflavin biosynthesis relies on the purine pathway, because GTP is the immediate precursor. Accordingly, it seems reasonably that redirecting metabolic flux toward purine biosynthesis might improve riboflavin production. In fact, previous studies described that supplementation with riboflavin precursors can enhance the production of the vitamin in A. gossypii cultures [3, 31]. PRPP is an essential metabolite in the de novo and salvage purine pathways and it has been reported that a considerable fraction of the intracellular pool of PRPP is consumed by the purine pathways [7]. We therefore explored the effect of increasing the activity of PRPP synthetase on riboflavin production in A. gossypii.
AGL080C gene is a syntenic ortholog of PRS3 from S. cerevisiae and their gene products share 88% of the amino acids. On the other hand, AGR371C shows a high degree of similarity with both PRS2 and PRS4 from S. cerevisiae and displays 80% of identity at protein level. Furthermore, AGR371C is flanked by the UBP9 and UBC6 genes, thus exhibiting a high level of synteny with regard to the genomic locations of PRS2 and PRS4 in S. cerevisiae. This redundancy between PRS genes and their chromosomal synteny supports the idea that a genome duplication event of ancestral yeast genome would have occurred to originate the S. cerevisiae genome [32].
The homology between PRS genes from A. gossypii and S. cerevisiae together with our results concerning the AgPRS2,4 and AgPRS3 disruptions indicate that two PRPP synthetase interaction complexes may also exist in A. gossypii. We found three possible phenotypes when AgPRS2,4 and AgPRS3 disruptions were combined: synthetic lethality for the double mutant, a growth alteration and an impairment in PRPP synthetase activity for the Δprs3 strain, and a slight reduction in total Prs activity that did not elicit phenotypic alterations in the Δprs2,4 mutant. Thus, similarly to what occurs in yeast it could be speculated that in A. gossypii Prs2,4 and Prs3 function as two different PRPP synthetase units that interact with Prs5 and Prs1, respectively, to yield physiologically specialized enzymatic complexes. The different phenotypes obtained with AgPRS2,4 and AgPRS3 disruptions can be explained in terms of a different contribution of each isoform to the specific PRPP synthetase functional unit.
Another important issue concerning AgPRS3 disruption is that the Δprs3 strain displayed an abnormal highly branched growth pattern. Previous work has described that Prs gene products are able to interact with a number of proteins and to affect different biological processes and, in particular, cell integrity signaling through Rlm1. As a consequence, perturbations in the expression of PRS genes may result in many unexpected cellular events, such as an abnormal increased chitin content and other cell wall alterations [33–35]. It has also been described that the mechanisms controlling cell polarity, and therefore hyphal morphogenesis, are fairly homologous between S. cerevisiae and A. gossypii [36]. It therefore seems reasonable to posit that alterations in cell polarity and cell wall morphogenesis might affect the branching pattern of the A. gossypii hyphae.
As mentioned above, the altered branching pattern of the Δprs3 strain could account for the condensed phenotype of the colonies that finally resulted in a slightly higher biomass. In contrast, the opposite effect has been reported when either AgPRS2,4 or agprs2,4-IQ alleles are overexpressed in Arabidopsis thaliana and Nicotiana tabacum [37], demonstrating that variations in the PRPP synthetase activity may have pleiotropic effects. Perhaps the occurrence and interaction of fairly heterologous isoforms of PRPP synthetases from A. gossypii together with endogenous isoforms of plant origin or other proteins in the plant cell could provide an explanation for the effect on biomass accumulation.
Our results demonstrate that the PRPP synthetases from A. gossypii are inhibited by ADP. The abolishment of ADP inhibition in A. gossypii and overexpression of PRS genes result in: i) an increase in PRPP synthetase activity and ii) improved riboflavin production, which was the main objective of the present work. However, unlike the overexpression of PRS3 alleles, the increase in the levels of either PRS2,4 or prs2,4-IQ mRNA was not correlated with a marked rise in PRPP synthetase activity, confirming that PRS2,4 and PRS3 contribute unequally to total Prs activity. However, an improvement of 80% in the production of riboflavin was obtained in the PRS-engineered strains, which is clearly of significance as regards biotechnological endeavors. Nonetheless, we failed to find a significant enhancement in the production of the vitamin when the prs-IQ alleles were expressed. This may be explained in terms of the existence of a strict transcriptional and metabolic regulation of the purine pathway downstream from the synthesis of PRPP, reported previously in A. gossypii and S. cerevisiae [3, 4, 38–42]. In addition, it is also to be expected that the intracellular concentration of ADP in the PRS-overexpressing strains would be insufficient to cause inhibition and this would therefore account for the analogous behaviour of the overexpressing mutant strains as compared with the wild-type overexpressing strains.
Conclusion
In this study we have demonstrated that partially increasing enzymatic PRPP synthetase activity results directly in enhanced riboflavin overproduction in A. gossypii. We have engineered A. gossypii strains that show a significant improvement in vitamin production. In terms of industrial production, a 80% increase in the fermentation process represents an extremely relevant advance. Further manipulations might be performed to entirely deregulate the purine pathway and to increase GTP availability and hence riboflavin production.
We also show that different mutimeric interacting PRPP synthetase complexes may exist in A. gossypii, as described for the S. cerevisiae enzymes. Additionally, we prove that modifying PRPP synthetase activity in A. gossypii affects its growth pattern, suggesting that homeostasis of the PRPP intracellular pool must be important for the cell integrity and mycelial growth of A. gossypii.
References
Vandamme EJ: Production of vitamins, coenzymes and related biochemicals by biotechnological processes. J Chem Technol Biotechnol. 1992, 53 (4): 313-327.
Stahmann KP, Arst HN, Althofer H, Revuelta JL, Monschau N, Schlupen C, Gatgens C, Wiesenburg A, Schlosser T: Riboflavin, overproduced during sporulation of Ashbya gossypii, protects its hyaline spores against ultraviolet light. Environ Microbiol. 2001, 3 (9): 545-550. 10.1046/j.1462-2920.2001.00225.x.
Jimenez A, Santos MA, Pompejus M, Revuelta JL: Metabolic engineering of the purine pathway for riboflavin production in Ashbya gossypii. Applied and Environmental Microbiology. 2005, 71 (10): 5743-5751. 10.1128/AEM.71.10.5743-5751.2005.
Mateos L, Jimenez A, Revuelta JL, Santos MA: Purine biosynthesis, riboflavin production, and trophic-phase span are controlled by a Myb-related transcription factor in the fungus Ashbya gossypii. Appl Environ Microbiol. 2006, 72 (7): 5052-5060. 10.1128/AEM.00424-06.
Revuelta JL, Buitrago MJ, Santos MA: Riboflavin-biosynthesis in fungi. Patent nr. WO9526406. In., C12N15/52 edn: BASF AG (DE). 1998
Rolfes RJ: Regulation of purine nucleotide biosynthesis: in yeast and beyond. Biochem Soc Trans. 2006, 34 (Pt 5): 786-790.
Hove-Jensen B: Mutation in the phosphoribosylpyrophosphate synthetase gene (prs) that results in simultaneous requirements for purine and pyrimidine nucleosides, nicotinamide nucleotide, histidine, and tryptophan in Escherichia coli. J Bacteriol. 1988, 170 (3): 1148-1152.
Becker MA, Puig JG, Mateos FA, Jimenez ML, Kim M, Simmonds HA: Neurodevelopmental impairment and deranged PRPP and purine nucleotide synthesis in inherited superactivity of PRPP synthetase. Adv Exp Med Biol. 1989, 253A: 15-22.
Becker MA, Smith PR, Taylor W, Mustafi R, Switzer RL: The genetic and functional basis of purine nucleotide feedback-resistant phosphoribosylpyrophosphate synthetase superactivity. J Clin Invest. 1995, 96 (5): 2133-2141. 10.1172/JCI118267.
de Brouwer AP, Williams KL, Duley JA, van Kuilenburg AB, Nabuurs SB, Egmont-Petersen M, Lugtenberg D, Zoetekouw L, Banning MJ, Roeffen M, Hamel BC, Weaving L, Ouvrier RA, Donald JA, Wevers RA, Christodoulou J, van Bokhoven H: Arts syndrome is caused by loss-of-function mutations in PRPS1. Am J Hum Genet. 2007, 81 (3): 507-518. 10.1086/520706.
Kim HJ, Sohn KM, Shy ME, Krajewski KM, Hwang M, Park JH, Jang SY, Won HH, Choi BO, Hong SH, Kim BJ, Suh YL, Ki CS, Lee SY, Kim SH, Kim JW: Mutations in PRPS1, which encodes the phosphoribosyl pyrophosphate synthetase enzyme critical for nucleotide biosynthesis, cause hereditary peripheral neuropathy with hearing loss and optic neuropathy (cmtx5). Am J Hum Genet. 2007, 81 (3): 552-558. 10.1086/519529.
Gattiker A, Rischatsch R, Demougin P, Voegeli S, Dietrich FS, Philippsen P, Primig M: Ashbya Genome Database 3.0: a cross-species genome and transcriptome browser for yeast biologists. BMC Genomics. 2007, 8: 9-10.1186/1471-2164-8-9.
Carter AT, Narbad A, Pearson BM, Beck KF, Logghe M, Contreras R, Schweizer M: Phosphoribosylpyrophosphate synthetase (PRS): a new gene family in Saccharomyces cerevisiae. Yeast. 1994, 10 (8): 1031-1044. 10.1002/yea.320100805.
Hernando Y, Parr A, Schweizer M: PRS5, the fifth member of the phosphoribosyl pyrophosphate synthetase gene family in Saccharomyces cerevisiae, is essential for cell viability in the absence of either PRS1 or PRS3. J Bacteriol. 1998, 180 (23): 6404-6407.
Hernando Y, Carter AT, Parr A, Hove-Jensen B, Schweizer M: Genetic analysis and enzyme activity suggest the existence of more than one minimal functional unit capable of synthesizing phosphoribosyl pyrophosphate in Saccharomyces cerevisiae. J Biol Chem. 1999, 274 (18): 12480-12487. 10.1074/jbc.274.18.12480.
Fox IH, Kelley WN: Human phosphoribosylpyrophosphate synthetase. Distribution, purification, and properties. J Biol Chem. 1971, 246 (18): 5739-5748.
Hove-Jensen B: Heterooligomeric phosphoribosyl diphosphate synthase of Saccharomyces cerevisiae: combinatorial expression of the five PRS genes in Escherichia coli. J Biol Chem. 2004, 279 (39): 40345-40350. 10.1074/jbc.M405480200.
Hove-Jensen B, Harlow KW, King CJ, Switzer RL: Phosphoribosylpyrophosphate synthetase of Escherichia coli. Properties of the purified enzyme and primary structure of the prs gene. J Biol Chem. 1986, 261 (15): 6765-6771.
Krath BN, Eriksen TA, Poulsen TS, Hove-Jensen B: Cloning and sequencing of cDNAs specifying a novel class of phosphoribosyl diphosphate synthase in Arabidopsis thaliana. Biochim Biophys Acta. 1999, 1430 (2): 403-408.
Kadziola A, Jepsen CH, Johansson E, McGuire J, Larsen S, Hove-Jensen B: Novel class III phosphoribosyl diphosphate synthase: structure and properties of the tetrameric, phosphate-activated, non-allosterically inhibited enzyme from Methanocaldococcus jannaschii. J Mol Biol. 2005, 354 (4): 815-828. 10.1016/j.jmb.2005.10.001.
Li S, Lu Y, Peng B, Ding J: Crystal structure of human phosphoribosylpyrophosphate synthetase 1 reveals a novel allosteric site. Biochem J. 2007, 401 (1): 39-47. 10.1042/BJ20061066.
Forster C, Santos MA, Ruffert S, Kramer R, Revuelta JL: Physiological consequence of disruption of the VMA1 gene in the riboflavin overproducer Ashbya gossypii. J Biol Chem. 1999, 274 (14): 9442-9448. 10.1074/jbc.274.14.9442.
Sherman F, Fink GR, Hicks J: Methods in Yeast Genetics. 1986, Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press
Schlupen C, Santos MA, Weber U, de Graaf A, Revuelta JL, Stahmann KP: Disruption of the SHM2 gene, encoding one of two serine hydroxymethyltransferase isoenzymes, reduces the flux from glycine to serine in Ashbya gossypii. Biochem J. 2003, 369 (Pt 2): 263-273. 10.1042/BJ20021224.
Santos MA, Mateos L, Stahmann KP, Revuelta JL: Insertional Mutagenesis in the Vitamin B2 Producer Fungus Ashbya gossypii. Methods in Biotechnology. 2004, Totowa, New Jersey: Humana Press Inc, 18:
Steiner S, Philippsen P: Sequence and promoter analysis of the highly expressed TEF gene of the filamentous fungus Ashbya gossypii. Mol Gen Genet. 1994, 242 (3): 263-271. 10.1007/BF00280415.
Jensen KF, Houlberg U, Nygaard P: Thin-layer chromatographic methods to isolate 32P-labeled 5-phosphoribosyl-alpha-1-pyrophosphate (PRPP): determination of cellular PRPP pools and assay of PRPP synthetase activity. Anal Biochem. 1979, 98 (2): 254-263. 10.1016/0003-2697(79)90138-6.
Stahmann KP, Revuelta JL, Seulberger H: Three biotechnical processes using Ashbya gossypii, Candida famata, or Bacillus subtilis compete with chemical riboflavin production. Appl Microbiol Biotechnol. 2000, 53 (5): 509-516. 10.1007/s002530051649.
Eriksen TA, Kadziola A, Bentsen AK, Harlow KW, Larsen S: Structural basis for the function of Bacillus subtilis phosphoribosyl-pyrophosphate synthetase. Nat Struct Biol. 2000, 7 (4): 303-308. 10.1038/74069.
Wendland J, Ayad-Durieux Y, Knechtle P, Rebischung C, Philippsen P: PCR-based gene targeting in the filamentous fungus Ashbya gossypii. Gene. 2000, 242 (1–2): 381-391. 10.1016/S0378-1119(99)00509-0.
Mac LJ: The effects of certain purines and pyrimidines upon the production of riboflavin by Eremothecium ashbyii. J Bacteriol. 1952, 63 (2): 233-241.
Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, Louis EJ, Mewes HW, Murakami Y, Philippsen P, Tettelin H, Oliver SG.: Life with 6000 genes. Science. 1996, 274 (5287): 563-547. 10.1126/science.274.5287.546.
Vavassori S, Wang K, Schweizer LM, Schweizer M: Ramifications of impaired PRPP synthesis in Saccharomyces cerevisiae. Biochem Soc Trans. 2005, 33 (Pt 6): 1418-1420.
Vavassori S, Wang K, Schweizer LM, Schweizer M: In Saccharomyces cerevisiae, impaired PRPP synthesis is accompanied by valproate and Li+ sensitivity. Biochem Soc Trans. 2005, 33 (Pt 5): 1154-1157.
Wang K, Vavassori S, Schweizer LM, Schweizer M: Impaired PRPP-synthesizing capacity compromises cell integrity signalling in Saccharomyces cerevisiae. Microbiology. 2004, 150 (Pt 10): 3327-3339. 10.1099/mic.0.27373-0.
Philippsen P, Kaufmann A, Schmitz HP: Homologues of yeast polarity genes control the development of multinucleated hyphae in Ashbya gossypii. Curr Opin Microbiol. 2005, 8 (4): 370-377. 10.1016/j.mib.2005.06.021.
Koslowsky S, Riegler H, Bergmuller E, Zrenner R: Higher biomass accumulation by increasing phosphoribosylpyrophosphate synthetase activity in Arabidopsis thaliana and Nicotiana tabacum. Plant Biotechnol J. 2007
Daignan-Fornier B, Fink GR: Coregulation of purine and histidine biosynthesis by the transcriptional activators BAS1 and BAS2. Proc Natl Acad Sci USA. 1992, 89 (15): 6746-6750. 10.1073/pnas.89.15.6746.
Escobar-Henriques M, Daignan-Fornier B: Transcriptional regulation of the yeast gmp synthesis pathway by its end products. J Biol Chem. 2001, 276 (2): 1523-1530. 10.1074/jbc.M007926200.
Holmes EW, McDonald JA, McCord JM, Wyngaarden JB, Kelley WN: Human glutamine phosphoribosylpyrophosphate amidotransferase. Kinetic and regulatory properties. J Biol Chem. 1973, 248 (1): 144-150.
Holmes EW, Pehlke DM, Kelley WN: Human IMP dehydrogenase. Kinetics and regulatory properties. Biochim Biophys Acta. 1974, 364 (2): 209-217.
Weyden Van der MB, Kelly WN: Human adenylosuccinate synthetase. Partial purification, kinetic and regulatory properties of the enzyme from placenta. J Biol Chem. 1974, 249 (22): 7282-7289.
Acknowledgements
This work was supported in part by BASF AG and grant AGL2005-07245-C03-03 from the Ministerio de Educación y Ciencia, Spain. We thank F. Martín for excellent technical help, and N. Skinner for correcting the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
JLR conceived the pivotal idea of the study. JLR and MAS co-designed the experiments and supervised the work. AJ performed the experiments and wrote the manuscript. All authors have read and approved the final version of the manuscript.
Electronic supplementary material
12896_2008_336_MOESM2_ESM.pdf
Additional file 2: Alignments of PRS proteins. Underlined in red are the residues involved in ribose-5-phosphate binding; underlined in blue are the residues involved in pyrophosphate binding. Red circles indicate residues that have been described to participate in catalysis in the B. subtilis, E. coli and human PRPP synthetases. (PDF 784 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Jiménez, A., Santos, M.A. & Revuelta, J.L. Phosphoribosyl pyrophosphate synthetase activity affects growth and riboflavin production in Ashbya gossypii. BMC Biotechnol 8, 67 (2008). https://doi.org/10.1186/1472-6750-8-67
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1472-6750-8-67