Abstract
The cellulolytic myxobacterium Sorangium cellulosum is able to efficiently degrade many kinds of polysaccharides, but none of the enzymes involved have been characterized. In this paper, a xylanase gene (xynA) was cloned from S. cellulosum So9733-1 using thermal asymmetric interlaced PCR. The gene is composed of 1,209 bp and has only 52.27% G + C content, which is much lower than that of most myxobacterial DNA reported (67–72%). Gene xynA encodes a 402 amino acid protein that contains a single catalytic domain belonging to the glycoside hydrolase family 10. The novel xylanase gene, xynA, was expressed in Escherichia coli BL21 (DE3) and the recombinant protein (r-XynA) was purified by Ni-affinity chromatography. The r-XynA had the optimum temperature of 30–35°C and exhibited 33.3% activity at 5°C and 13.7% activity at 0°C. Approximately 80% activity was lost after 20-min pre-incubation at 50°C. These results indicate that r-XynA is a cold-active xylanase with low thermostability. At 30°C, the K m values of r-XynA on beechwood xylan, birchwood xylan, and oat spelt xylan were 25.77 ± 4.16, 26.52 ± 4.78, and 38.13 ± 5.35 mg/mL, respectively. The purified r-XynA displayed optimum activity at pH 7.0. The activity of r-XynA was enhanced by the presence of Ca2+. The r-XynA hydrolyzed beechwood xylan, birchwood xylan, and xylooligosaccharides (xylotriose, xylotetraose, and xylopentose) to produce primarily xylose and xylobiose. To our knowledge, this is the first report on the characterization of a xylanase from S. cellulosum.



Similar content being viewed by others
References
Beg QK, Kapoor M, Mahajan L, Hoondal GS (2001) Microbial xylanases and their industrial applications: a review. Appl Microbiol Biotechnol 56:326–338
Bhat MK (2000) Cellulases and related enzymes in biotechnology. Biotechnol Adv 18:355–383
Biely P (1985) Microbial xylanolytic systems. Trends Biotechnol 3:286–290
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Collins T, Meuwis MA, Stals I, Claeyssens M, Feller G, Gerday C (2002) A novel family 8 xylanase, functional and physicochemical characterization. J Biol Chem 277:35133–35139
Collins T, Gerday C, Feller G (2005) Xylanases, xylanase families and extremophilic xylanases. FEMS Microbiol Rev 29:3–23
Collins T, Roulling F, Piette F, Marx JC, Feller G, Gerday C, D’Amico S (2008) Fundamentals of cold-adapted enzymes. In: Margesin R, Schinner F, Marx JC, Gerday C (eds) Psychrophiles: from biodiversity to biotechnology. Springer, Berlin, pp 211–227
Ebringerová A, Heinze T (2000) Xylan and xylan derivatives—biopolymers with valuable properties, 1. Naturally occurring xylans structures, isolation procedures and properties. Macromol Rapid Commun 21:542–556
Gerday C, Aittaleb M, Bentahir M, Chessa JP, Claverie P, Collins T, D’Amico S, Dumont J, Garsoux G, Georlette D, Hoyoux A, Lonhienne T, Meuwis MA, Feller G (2000) Cold-adapted enzymes: from fundamentals to biotechnology. Trends Biotechnol 18:103–107
Gerth K, Pradella S, Perlova O, Beyer S, Müller R (2003) Myxobacteria: proficient producers of novel natural products with various biological activities-past and future biotechnological aspects with the focus on the genus Sorangium. J Biotechnol 106:233–253
Guo B, Chen XL, Sun CY, Zhou BC, Zhang YZ (2009) Gene cloning, expression and characterization of a new cold-active and salt-tolerant endo-β-1,4-xylanase from marine Glaciecola mesophila KMM 241. Appl Microbiol Biotechnol 84:1107–1115
Haegeman A, Vanholme B, Gheysen G (2009) Characterization of a putative endoxylanase in the migratory plant-parasitic nematode Radopholus similis. Mol Plant Pathol 10:389–401
Hou PB, Li YZ, Wu BH, Yan ZC, Yan BX, Gao PJ (2006a) Cellulolytic complex exists in cellulolytic myxobacterium Sorangium. Enzyme Microb Technol 38:273–278
Hou YH, Wang TH, Long H, Zhu HY (2006b) Novel cold-adaptive Penicillium strain FS010 secreting thermo-labile xylanase isolated from Yellow Sea. Acta Biochim Biophys Sin 38:142–149
Khandeparker R, Numan M (2008) Bifunctional xylanases and their potential use in biotechnology. J Ind Microbiol Biotechnol 35:635–644
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Lee CC, Kibblewhite-Accinelli RE, Wagschal K, Robertson GH, Wong DW (2006a) Cloning and characterization of a cold-active xylanase enzyme from an environmental DNA library. Extremophiles 10:295–300
Lee CC, Smith M, Kibblewhite-Accinelli RE, Williams TG, Wagschal K, Robertson GH, Wong DW (2006b) Isolation and characterization of a cold-active xylanase enzyme from Flavobacterium sp. Curr Microbiol 52:112–116
Liu YG, Whittier RF (1995) Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25:674–681
Luo H, Yang J, Li J, Shi P, Huang H, Bai Y, Fan Y, Yao B (2010) Molecular cloning and characterization of the novel acidic xylanase XYLD from Bispora sp. MEY-1 that is homologous to family 30 glycosyl hydrolases. Appl Microbiol Biotechnol 86:1829–1839
Lv Z, Yang J, Yuan H (2008) Production, purification and characterization of an alkaliphilic endo-β-1,4-xylanase from a microbial community EMSD5. Enzyme Microb Technol 43:343–348
Miller GL, Blum R, Glennon WE, Burton AL (1960) Measurement of carboxymethylcellulase activity. Anal Biochem 1:127–132
Petrescu I, Lamotte-Brasseur J, Chessa JP, Ntarima P, Claeyssens M, Devreese B, Marino G, Gerday C (2000) Xylanase from the psychrophilic yeast Cryptococcus adeliae. Extremophiles 4:137–144
Pollet A, Delcour JA, Courtin CM (2010) Structural determinants of the substrate specificities of xylanases from different glycoside hydrolase families. Crit Rev Biotechnol 30:176–191
Salles BC, Cunha RB, Fontes W, Sousa MV, Filho EX (2000) Purification and characterization of a new xylanase from Acrophialophora nainiana. J Biotechnol 81:199–204
Schneiker S, Perlova O, Kaiser O, Gerth K, Alici A, Altmeyer MO, Bartels D, Beke T, Beyer S, Bode E, Bode HB, Bolten CJ, Choudhuri JV, Doss S, Elnakady YA, Frank B, Gaigalat L, Goesmann A, Groeger C, Gross F, Jelsbak L, Jelsbak L, Kalinowski J, Kegler C, Knauber T, Konietzny S, Kopp M, Krause L, Krug D, Linke B, Mahmud T, Martinez-Arias R, McHardy AC, Merai M, Meyer F, Mormann S, Muñoz-Dorado J, Perez J, Pradella S, Rachid S, Raddatz G, Rosenau F, Rückert C, Sasse F, Scharfe M, Schuster SC, Suen G, Treuner-Lange A, Velicer GJ, Vorhölter F-J, Weissman KJ, Welch RD, Wenzel SC, Whitworth DE, Wilhelm S, Wittmann C, Blöcker H, Pühler A, Müller R (2007) Complete genome sequence of the myxobacterium Sorangium cellulosum. Nat Biotechnol 25:1281–1289
Shallom D, Shoham Y (2003) Microbial hemicellulases. Curr Opin Microbiol 6:219–228
Shimkets LJ, Dworkin M, Reichenbach H (2006) The myxobacteria. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E (eds) The prokaryotes. Springer, New York, pp 31–115
Solomon V, Teplitsky A, Shulami S, Zolotnitsky G, Shoham Y, Shoham G (2007) Structure-specificity relationships of an intracellular xylanase from Geobacillus stearothermophilus. Acta Crystallogr D Biol Crystallogr 63:845–859
Spiwok V, Lipovová P, Skálová T, Dušková J, Dohnálek J, Hašek J, Russell NJ, Králová B (2007) Cold-active enzymes studied by comparative molecular dynamics simulation. J Mol Model 13:485–497
St John FJ, Gonzalez JM, Pozharski E (2010) Consolidation of glycosyl hydrolase family 30: a dual domain 4/7 hydrolase family consisting of two structurally distinct groups. FEBS Lett 584:4435–4441
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Turkiewicz M, Kalinowska H, Zielińska M, Bielecki S (2000) Purification and characterization of two endo-1,4-β-xylanases from Antarctic krill, Euphausia superba Dana. Comp Biochem Physiol B Biochem Mol Biol 127:325–335
Wang G, Luo H, Wang Y, Huang H, Shi P, Yang P, Meng K, Bai Y, Yao B (2011) A novel cold-active xylanase gene from the environmental DNA of goat rumen contents: direct cloning, expression and enzyme characterization. Bioresour Technol 102:3330–3336
Yan ZC, Wang B, Li YZ, Gong X, Zhang HQ, Gao PJ (2003) Morphologies and phylogenetic classification of cellulolytic myxobacteria. Syst Appl Microbiol 26:104–109
Zhang M, Jiang Z, Yang S, Hua C, Li L (2010) Cloning and expression of a Paecilomyces thermophila xylanase gene in E. coli and characterization of the recombinant xylanase. Bioresour Technol 101:688–695
Zhou J, Huang H, Meng K, Shi P, Wang Y, Luo H, Yang P, Bai Y, Zhou Z, Yao B (2009) Molecular and biochemical characterization of a novel xylanase from the symbiotic Sphingobacterium sp. TN19. Appl Microbiol Biotechnol 85:323–333
Acknowledgments
This work was financially supported by grants from the National Natural Science Foundation of China (30670028) and the Shandong Province Natural Science Foundation (ZR2009DM016).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
A phylogenetic tree constructed using the neighbor-joining method (MEGA 4.1). Bootstrap values (n = 1,000 replicates) are reported as percentages. The scale bar represents the number of changes per amino acid position. Accession numbers and optimum temperatures of the xylanases are given at the end of each species name (JPEG 37 kb)
Fig. S2
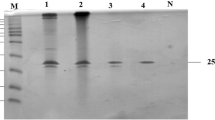
SDS-PAGE analysis of the expression and purification of the r-XynA. M, protein molecular mass marker; lane 1, soluble cell lysates of induced E. coli BL21 (DE3) carrying pET-22b(+); lane 2, soluble cell lysates of uninduced E. coli BL21 (DE3) carrying pET-xynA; lane 3, soluble cell lysates of induced E. coli BL21 (DE3) carrying pET-xynA; lane 4, purified r-XynA after Ni-affinity (JPEG 12 kb)
Rights and permissions
About this article
Cite this article
Wang, SY., Hu, W., Lin, XY. et al. A novel cold-active xylanase from the cellulolytic myxobacterium Sorangium cellulosum So9733-1: gene cloning, expression, and enzymatic characterization. Appl Microbiol Biotechnol 93, 1503–1512 (2012). https://doi.org/10.1007/s00253-011-3480-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-011-3480-3