Abstract
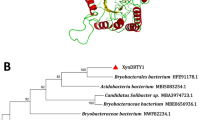
A xylanase-encoding gene, designated xynA19, was cloned from Sphingobacterium sp. TN19—a symbiotic bacterium isolated from the gut of Batocera horsfieldi larvae—and expressed in Escherichia coli BL21 (DE3). The full-length xynA19 (1,155 bp in length) encodes a 384-residue polypeptide (XynA19) containing a predicted signal peptide of 24 residues and a catalytic domain belonging to glycosyl hydrolase family 10 (GH 10). The deduced amino acid sequence of XynA19 is most similar (53.1% identity) to an endo-1,4-β-xylanase from Prevotella bryantii B14. Phylogenetic analysis of GH 10 Bacteroidia xylanases indicated that GH 10 xylanases from Sphingobacteria were separated into two clusters, and XynA19 is more closely related to the xylanases of Bacteroidia from gut or rumen than to those of Flavobacteria and Sphingobacteria from other sources. Recombinant XynA19 (r-XynA19) showed apparent optimal activity at pH 6.5 and 45°C. Compared with thermophilic and mesophilic counterparts, r-XynA19 was more active at low temperatures, retaining >65% of its maximum activity at 20–28°C and ~40% even at 10°C, and modeling indicated that XynA19 has fewer hydrogen bonds and salt bridges. These properties suggest that XynA19 has various potential applications, especially in aquaculture and the food industry.






Similar content being viewed by others
References
Akila G, Chandra TS (2003) A novel cold-tolerant Clostridium strain PXYL1 isolated from a psychrophilic cattle manure digester that secretes thermolabile xylanase and cellulase. FEMS Microbiol Lett 219:63–67
Breznak JA, Brune A (1994) Role of microorganisms in the digestion of lignocellulose by termites. Annu Rev Entomol 39:453–487
Cao L, Wang W, Yang C, Yang Y, Dianac J, Yakupitiyage A, Luo Z, Li D (2007) Application of microbial phytase in fish feed. Enzyme Microb Technol 40:497–507
Ceroni A, Passerini A, Vullo A, Frasconi P (2006) DISULFIND: a disulfide bonding state and cysteine connectivity prediction server. Nucleic Acids Res 34:W177–W181
Collins T, Meuwis MA, Stals I, Claeyssens M, Feller G, Gerday C (2002) A novel family 8 xylanase, functional and physicochemical characterization. J Biol Chem 277:35133–35139
Collins T, Gerday C, Feller G (2005) Xylanases, xylanase families and extremophilic xylanases. FEMS Microbiol Rev 29:3–23
Dehority BA (1969) Pectin-fermenting bacteria isolated from the bovine rumen. J Bacteriol 99:189–196
Eisenberg D, Lüthy R, Bowie JU (1997) VERIFY3D: assessment of protein models with three-dimensional profiles. Methods Enzymol 277:396–404
Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, Bateman A (2008) The Pfam protein families database. Nucleic Acids Res 36:D281–D288
Gasparic A, Marinsek-Logar R, Martin J, Wallace RJ, Nekrep FV, Flint HJ (1995a) Isolation of genes encoding β-D-xylanase, β-D-xylosidase and α-L-arabinofuranosidase activities from the rumen bacterium Prevotella ruminicola B14. FEMS Microbiol Lett 125:135–141
Gasparic A, Martin J, Daniel AS, Flint HJ (1995b) A xylan hydrolase gene cluster in Prevotella ruminicola B14: sequence relationships, synergistic interactions, and oxygen sensitivity of a novel enzyme with exoxylanase and beta-(1, 4)-xylosidase activities. Appl Environ Microbiol 61:2958–2964
Gomes J, Gomes I, Steiner W (2000) Thermolabile xylanase of the Antarctic yeast Cryptococcus adeliae: production and properties. Extremophiles 4:227–235
Hayashi H, Abe T, Sakamoto M, Ohara H, Ikemura T, Sakka K, Benno Y (2005) Direct cloning of genes encoding novel xylanases from the human gut. Can J Microbiol 51:251–259
Heo S, Kwak J, Oh HW, Park DS, Bae KS, Shin DH, Park HY (2006) Characterization of an extracellular xylanase in Paenibacillus sp. HY-8 isolated from an herbivorous longicorn beetle. J Microbiol Biotechnol 16:1753–1759
Humphrey W, Dalke A, Schulten K (1996) VMD—visual molecular dynamics. J Mol Graph 14:33–38
Ihsanawati KT, Kaneko T, Morokuma C, Yatsunami R, Sato T, Nakamura S, Tanaka N (2005) Structural basis of the substrate subsite and the highly thermal stability of xylanase 10B from Thermotoga maritima MSB8. Proteins 61:999–1009
Jiang ZQ, Kobayashi A, Ahsan MM, Lite L, Kitaoka M, Hayashi K (2001) Characterization of a thermostable family 10 endo-xylanase (XynB) from Thermotoga maritima that cleaves p-nitrophenyl-β-D-xyloside. J Biosci Bioeng 92:423–428
Khasin A, Alchanati I, Shoham Y (1993) Purification and characterization of a thermostable xylanase from Bacillus stearothermophilus T-6. Appl Environ Microbiol 59:1725–1730
Lee CC, Kibblewhite-Accinelli RE, Wagschal K, Robertson GH, Wong DW (2006a) Cloning and characterization of a cold-active xylanase enzyme from an environmental DNA library. Extremophiles 10:295–300
Lee CC, Smith M, Kibblewhite-Accinelli RE, Williams TG, Wagschal K, Robertson GH, Wong DW (2006b) Isolation and characterization of a cold-active xylanase enzyme from Flavobacterium sp. Curr Microbiol 52:112–116
Li N, Meng K, Wang Y, Shi P, Luo H, Bai Y, Yang P, Yao B (2008) Cloning, expression, and characterization of a new xylanase with broad temperature adaptability from Streptomyces sp. S9. Appl Microbiol Biotechnol 80:231–240
Li N, Shi P, Yang P, Wang Y, Luo H, Bai Y, Zhou Z, Yao B (2009) Cloning, expression, and characterization of a new Streptomyces sp. S27 xylanase for which xylobiose is the main hydrolysis product. Appl Microbiol Biotechnol doi: https://doi.org/10.1007/s1201
Liu YG, Whittier RF (1995) Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25:674–681
Liu JR, Duan CH, Zhao X, Tzen JT, Cheng KJ, Pai CK (2008) Cloning of a rumen fungal xylanase gene and purification of the recombinant enzyme via artificial oil bodies. Appl Microbiol Biotechnol 79:225–233
Manimaran A, Kumar KS, Permaul K, Singh S (2009) Hyper production of cellulase-free xylanase by Thermomyces lanuginosus SSBP on bagasse pulp and its application in biobleaching. Appl Microbiol Biotechnol 81:887–893
Miyazaki K, Miyamoto H, Mercer DK, Hirase T, Martin JC, Kojima Y, Flint HJ (2003) Involvement of the multidomain regulatory protein XynR in positive control of xylanase gene expression in the ruminal anaerobe Prevotella bryantii B14. J Bacteriol 185:2219–2226
Parinov S, Kondrichin I, Korzh V, Emelyanov A (2004) Tol2 transposon-mediated enhancer trap to identify developmentally regulated zebrafish genes in vivo. Dev Dyn 231:449–459
Park DS, Oh HW, Heo SY, Jeong WJ, Shin DH, Bae KS, Park HY (2007) Characterization of an extracellular lipase in Burkholderia sp. HY-10 isolated from a longicorn beetle. J Microbiol 45:409–417
Petrescu I, Lamotte-Brasseur J, Chessa JP, Ntarima P, Claeyssens M, Devreese B, Marino G, Gerday C (2000) Xylanase from the psychrophilic yeast Cryptococcus adeliae. Extremophiles 4:137–144
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera-a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612
Roy N, Rana MM, Uddin ATMS (2004) Isolation and some properties of new xylanase from the intestine of a herbivorous insect (Samia cynthia pryeri). J Biol Sci 4:27–33
Ruiz-Arribas A, Fernández-Abalos JM, Sánchez P, Garda AL, Santamariá RI (1995) Overproduction, purification, and biochemical characterization of a xylanase (Xys1) from Streptomyces halstedii JM8. Appl Environ Microbiol 61:2414–2419
Schloss PD, Delalibera I, Handelsman J, Raffa KF (2006) Bacteria associated with the guts of two wood-boring beetles: Anoplophora glabripennis and Saperda vestita (Cerambycidae). Environ Entomol 35:625–629
Silva Z, Horta C, da Costa MS, Chung AP, Rainey FA (2000) Polyphasic evidence for the reclassification of Rhodothermus obamensis Sako et al. 1996 as a member of the species Rhodothermus marinus Alfredsson et al. 1988. Int J Syst Evol Microbiol 50:1457–1461
Smith MT, Tobin PC, Bancroft J, Li GH, Gao RT (2004) Dispersal and spatiotemporal dynamics of Asian longhorned beetle (Coleoptera: Cerambycidae) in China. Environ Entomol 33:435–442
Subramaniyan S, Prema P (2002) Biotechnology of microbial xylanases: enzymology, molecular biology, and application. Crit Rev Biotechnol 22:33–64
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The Clustal X windows interface, flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
Turkiewicz M, Kalinowska H, Zielinśka M, Bielecki S (2000) Purification and characterization of two endo-1, 4-β-xylanases from Antarctic krill, Euphausia superba Dana. Comp Biochem Physiol B Biochem Mol Biol 127:325–335
van Petegem F, Collins T, Meuwis MA, Gerday C, Feller G, van Beeumen J (2003) The structure of a cold-adapted family 8 xylanase at 1.3 Å resolution. Structural adaptations to cold and investgation of the active site. J Biol Chem 278:7531–7539
Willard L, Ranjan A, Zhang H, Monzavi H, Boyko RF, Sykes BD, Wishart DS (2003) VADAR: a web server for quantitative evaluation of protein structure quality. Nucleic Acids Res 31:3316–3319
Xie G, Bruce DC, Challacombe JF, Chertkov O, Detter JC, Gilna P, Han CS, Lucas S, Misra M, Myers GL, Richardson P, Tapia R, Thayer N, Thompson LS, Brettin TS, Henrissat B, Wilson DB, McBride MJ (2007) Genome sequence of the cellulolytic gliding bacterium Cytophaga hutchinsonii. Appl Environ Microbiol 73:3536–3546
Acknowledgments
This work was supported by the National High Technology Research and Development Program of China (863 Program; No. 2007AA100601), the National Key Technology Research and Development Program of China (No. 2006BAD12B05-03), and the Chinese Program on Research for Public Good (Grant No. 2005DIB4J038). We are grateful to Professor Youqing Luo of Beijing Forestry University, China, for identification of Batocera horsfieldi.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhou, J., Huang, H., Meng, K. et al. Molecular and biochemical characterization of a novel xylanase from the symbiotic Sphingobacterium sp. TN19. Appl Microbiol Biotechnol 85, 323–333 (2009). https://doi.org/10.1007/s00253-009-2081-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-009-2081-x