Abstract
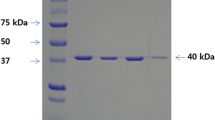
Bacillus subtilis endo-β-1,4-glucanase (Cel5A) hydrolyzes cellulose by cleavage of the internal bonds in the glucose chains, producing new ends randomly. Using directed evolution techniques of error-prone polymerase chain reaction (PCR) and DNA shuffling, several Cel5A variants with improved catalytic activity had been screened from the mutant library, which contained 71,000 colonies. Compared with the wild-type enzyme, the variants (M44-11, S75 and S78) showed 2.03 to 2.68-fold increased activities toward sodium carboxymethyl cellulose (CMC), while the M44-11 also exhibited a wider pH tolerance and higher thermostability. Structural models of M44-11, S75, S78, and WT proteins revealed that most of the substitutions were not located in the strictly conserved regions, except the mutation V255A of S75, which was closed to the nucleophile Glu257 in the catalytic center of the enzyme. Moreover, V74A and D272G of M44-11, which were not located in the substrate binding sites and the catalytic center, might result in improved stability and catalytic activity. These results provided useful references for directed evolution of the enzymes that belonged to the glycoside hydrolase family 5 (GH5).






Similar content being viewed by others
References
Arai T, Araki R, Tanaka A, Karita S, Kimura T, Sakka K, Ohmiya K (2003) Characterization of a cellulase containing a family 30 carbohydrate-binding module (CBM) derived from Clostridium thermocellum CelJ: Importance of the CBM to cellulose hydrolysis. J Bacteriol 185:504–512
Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Cao S, Liu Z, Guo A, Li Y, Zhang C, Gaobing W, Chunfang F, Tan Y, Chen H (2008) Efficient production and characterization of Bacillus anthracis lethal factor and a novel inactive mutant rLFm-Y236F. Protein Expr Purif 59:25–30
Castle LA, Siehl DL, Gorton R, Patten PA, Chen YH, Bertain S, Cho HJ, Duck N, Wong J, Liu DL, Lassner MW (2004) Discovery and directed evolution of a glyphosate tolerance gene. Science 304:1151–1154
Ducros V, Czjzek M, Belaich A, Gaudin C, Fierobe HP, Belaich JP, Davies GJ, Haser R (1995) Crystal structure of the catalytic domain of a bacterial cellulase belonging to family 5. Structure 3:939–949
Fan YH, Fang WG, Xiao YH, Yang XY, Zhang YJ, Bidochka MJ, Pei Y (2007) Directed evolution for increased chitinase activity. Appl Microbiol Biotechnol 76:135–139
Gloster TM, Macdonald JM, Tarling CA, Stick RV, Withers SG, Davies GJ (2004) Structural, thermodynamic, and kinetic analyses of tetrahydrooxazine-derived inhibitors bound to beta-glucosidases. J Biol Chem 279:49236–49242
Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723
Han SJ, Yoo YJ, Kang HS (1995) Characterization of a bifunctional cellulase and its structural gene—the cel gene of Bacillus sp. D04 has exoglucanase and endoglucanase activity. J Biol Chem 270:26012–26019
Henrissat B, Bairoch A (1996) Updating the sequence-based classification of glycosyl hydrolases. Biochem J 316:695–696
Kaper T, Brouns SJJ, Geerling ACM, De Vos WM, Van der Oost J (2002) DNA family shuffling of hyperthermostable beta-glycosidases. Biochem J 368:461–470
Kim MS, Lei XG (2008) Enhancing thermostability of Escherichia coli phytase AppA2 by error-prone PCR. Appl Microbiol Biotechnol 79:69–75
Kim JM, Kong IS, Yu JH (1987) Molecular cloning of an endoglucanase gene from an alkalophilic Bacillus sp. and its expression in Escherichia coli. Appl Environ Microbiol 53:2656–2659
Kim YS, Jung HC, Pan JG (2000) Bacterial cell surface display of an enzyme library for selective screening of improved cellulase variants. Appl Environ Microbiol 66:788–793
Leisola M, Turunen O (2007) Protein engineering: opportunities and challenges. Appl Microbiol Biotechnol 75:1225–1232
Li Y-H, Ding M, Wang J, Xu G-J, Zhao F (2006) A novel thermoacidophilic endoglucanase, Ba-EGA, from a new cellulose-degrading bacterium, Bacillus sp. AC-1. Appl Microbiol Biotechnol 70:430–436
Lo AC, MacKay RM, Seligy VL, Willick GE (1988) Bacillus subtilis beta-1,4-endoglucanase products from intact and truncated genes are secreted into the extracellular medium by Escherichia coli. Appl Environ Microbiol 54:2287–2292
McCarthy JK, Uzelac A, Davis DF, Eveleigh DE (2004) Improved catalytic efficiency and active site modification of 1,4-beta-d-glucan glucohydrolase A from Thermotoga neapolitana by directed evolution. J Biol Chem 279:11495–11502
Park JS, Nakamura A, Horinouchi S, Beppu T (1993) Identification of the cellulose-binding domain of a Bacillus subtilis endoglucanase distinct from its catalytic domain. Biosci Biotech Bioch 57:260–264
Percival Zhang YH, Himmel ME, Mielenz JR (2006) Outlook for cellulase improvement: screening and selection strategies. Biotechnol Adv 24:452–481
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual (3-volume set). Cold Spring Harbor Laboratory Press, Cold Spring Harbor New York
Schwede T, Kopp J, Guex N, Peitsch MC (2003) SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res 31:3381–3385
Shimon LJW, Pages S, Belaich A, Belaich J-P, Bayer EA, Lamed R, Shoham Y, Frolow F (2000) Structure of a family IIIa scaffoldin CBD from the cellulosome of Clostridium cellulolyticum at 2.2 ANG resolution. Acta Crystallogr D Biological Crystallography 56:1560–1568
Song JK, Rhee JS (2000) Simultaneous enhancement of thermostability and catalytic activity of phospholipase A1 by evolutionary molecular engineering. Appl and Environ Microbiol 66:890–894
van der Veen BA, Potocki-Veronese G, Albenne C, Joucla G, Monsan P, Remaud-Simeon M (2004) Combinatorial engineering to enhance amylosucrase performance: construction, selection, and screening of variant libraries for increased activity. Febs Lett 560:91–97
Varrot A, Davies GJ (2003) Direct experimental observation of the hydrogen-bonding network of a glycosidase along its reaction coordinate revealed by atomic resolution analyses of endoglucanase Cel5A. Acta Crystallogr D Biological Crystallography 59:447–452
Wang T, Liu X, Yu Q, Zhang X, Qu Y, Gao P, Wang T (2005) Directed evolution for engineering pH profile of endoglucanase III from Trichoderma reesei. Biomol Eng 22:89–94
Yang MJ, Jung SH, Shin ES, Kim J, Yun HD, Wong SL, Kim H (2004) Expression of a Bacillus subtilis endoglucanase in protease-deficient Bacillus subtilis strains. J Microbiol Biotechnol 14:430–434
Zhang S, Yin QY, Li YH, Ding M, Xu GJ, Zhao FK (2007) Molecular and biochemical characterization of Ba-EGA, a cellulase secreted by Bacillus sp. AC-1 from Ampullaria crosseans. Appl Microbiol Biotechnol 75:1327–1334
Zhao H, Arnold FH (1997) Optimization of DNA shuffling for high fidelity recombination. Nucleic Acids Res 25:1307–1308
Acknowledgment
We would like to thank Dr. Qifa Zhang for many valuable suggestions. This work was supported by grants from the National Natural Sciences Foundation of China (30770021 and 30570057) and the 111 Project (B07041).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lin, L., Meng, X., Liu, P. et al. Improved catalytic efficiency of Endo-β-1,4-glucanase from Bacillus subtilis BME-15 by directed evolution. Appl Microbiol Biotechnol 82, 671–679 (2009). https://doi.org/10.1007/s00253-008-1789-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-008-1789-3