Abstract
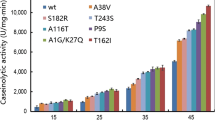
Directed evolution through DNA shuffling and screening was used to enhance the catalytic ability of a fungal, Beauveria bassiana, chitinase, Bbchit1. The Bbchit gene was first linked to various prokaryotic signal sequences and expressed in Escherichia coli. The signal peptide, PelB, from Erwinia carotovora resulted in greatest chitinase secretion into broth. The nucleotide sequence expressing PelB signal peptide was then incorporated into an E. coli vector to express Bbchit1 variants generated by three rounds of DNA shuffling. A Bbchit1 library with 150,000 variants was constructed with a nucleotide point mutation frequency of 0.6% and screened for chitinolytic activity. Two Bbchit1 variants (SHU-1 and SHU-2) were selected that showed increased chitinolytic activity compared to the wild type. Sequence analysis of these variants revealed mutations in amino acid residues that would not normally be considered for rational design of improved chitinase activity. The amino acid substitutions occurred outside of the two putative substrate-binding sites and the catalytic region.


Similar content being viewed by others
References
Arnold FH (1998) Design by directed evolution. Acc Chem Res 31:125–131
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Cristobal S, de Gier JW, Nielsen H, von Heijne G (1999) Competition between Sec-and TAT-dependent protein translocation in Escherichia coli. EMBO J 18:2982–2990
Fang W, Leng B, Xiao Y, Jin K, Ma J, Fan Y, Feng J, Yang X, Zhang Y, Pei Y (2005) Cloning of Beauveria bassiana chitinase gene Bbchit1 and its application to improve fungal strain virulence. Appl Environ Microbiol 71:363–370
Felse PA, Panda T (1999) Regulation and cloning of microbial chitinase genes. Appl Microbiol Biotechnol 51:141–151
Gooday GW (1999) Aggressive and defensive roles for chitinases. In: Jollės P, Muzzarelli RAA (eds) Chitin and chitinases. Birkhäuser Verlag, Basel, Switzerland, pp 157–169
Hinton JC, Sidebotham JM, Gill DR, Salmond GP (1989) Extracellular and periplasmic isoenzymes of pectate lyase from Erwinia carotovora subspecies carotovora belong to different gene families. Mol Microbiol 3:1785–1795
Inouye H, Barnes W, Beckwith J (1982) Signal sequence of alkaline phosphatase of Escherichia coli. J Bacteriol 149:434–439
Liu Z, SunZ, Leng Y (2006) Directed evolution and characterization of a novel d-pantonohydrolase from Fusarium moniliforme. J Agric Food Chem 54:5823–5830
Lorito M, Woo SL, Garcia I, Colucci G, Harman GE, Pintor-Toro JA, Filippone E, Muccifora S, Lawrence CB, Zoina A, Tuzun S, Scala F (1998) Genes from mycoparasitic fungi as a source for improving plant resistance to fungal pathogens. Proc Natl Acad Sci USA 95:7860–7865
Mauch F, Hadwiger LA, Boller T (1984) Ethylene: symptom, not signal for the induction of chitinase and β-1,3 glucanase in pea pods by pathogens and elicitors. Plant Physiol 76:607–611
Moore JC, Arnold FH (1996) Directed evolution of a para-nitrobenzyl esterase for aqueous-organic solvents. Nat Biotechnol 14:458–467
Moore GL, Maranas CD (2000) Modeling DNA mutation and recombination for directed evolution experiments. J Theor Biol 205:483–503
Nakamura K, Inouye M (1979) DNA sequence of the gene for the outer membrane lipoprotein of E. coli: an extremely AT-rich promoter. Cell 18:1109–1117
Oue S, Okamoto A, Yano T, Kagamiyama H (1999) Redesigning the substrate specificity of an enzyme by cumulative effects of the mutations of non-active site residues. J Biol Chem 274:2344–2349
Patil RS, Ghormade VV, Deshpande MV (2000) Chitinolytic enzymes: an exploration. Enzyme Microb Technol 26:473–483
Shimahara K, Takiguchi Y (1988) Preparation of crustacean chitin. Methods Enzymol 161:417–423
Stemmer WP (1994a) Rapid evolution of a protein in vitro by DNA shuffling. Nature 370:389–391
Stemmer WP (1994b) DNA shuffling by random fragmentation and reassembly: in vitro recombination for molecular evolution. Proc Natl Acad Sci USA 91:10747–10751
van der Veen BA, Potocki-Veronese G, Albenne C, Joucla G, Monsan P, Remaud-Simeon M (2004) Combinatorial engineering to enhance amylosucrase performance: construction, selection, and screening of variant libraries for increased activity. FEBS Lett 560:91–97
Wang SL, Hwang JR (2001) Microbial reclamation of shellfish wastes for the production of chitinases. Enzyme Microb Technol 28:376–382
Xu HF, Zhang XE, Zhang ZP, Zhang YM, Cass AEG (2003) Directed evolution of E. coli alkaline phosphatase towards higher catalytic activity. Biocatal Biotransform 21:41–47
Zhang JH, Dawes G, Stemmer WP (1997) Directed evolution of a fucosidase from a galactosidase by DNA shuffling and screening. Proc Natl Acad Sci USA 94:4504–4509
Acknowledgment
This work was supported by a China National Basic Research and Development Program grant (2003CB114203), a NSERC Discovery Grant to MJB, and a grant from the National Natural Sciences Foundation of China (30080001).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Fan, Y., Fang, W., Xiao, Y. et al. Directed evolution for increased chitinase activity. Appl Microbiol Biotechnol 76, 135–139 (2007). https://doi.org/10.1007/s00253-007-0996-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-007-0996-7