Abstract
Key message
Novel large-effect consistent QTL for anther extrusion (AE) to improve cross-pollination were mapped in doubled haploid populations derived from IPK gene bank spring wheat accessions. TaAP2-D, an ortholog of Cleistogamy1 in barley, is a likely candidate gene for AE in wheat.
Abstract
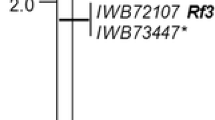
To establish a robust hybrid wheat breeding system, male lines harboring alleles that promote outcrossing should be developed. In this study, we developed two doubled haploid (DH) populations of hexaploid spring wheat (Triticum aestivum L.) by crossing accessions taken from IPK gene bank. In both populations, the phenotypic data of anther extrusion (AE) based on three years of field trials showed a wide variation and approximated a normal distribution. Both populations were genotyped with a 15 k Infinium single nucleotide polymorphism (SNP) array resulting in 3567 and 3457 polymorphic SNP markers for DH population-1 and DH population-2, respectively. Composite interval mapping identified quantitative trait loci (QTL) on chromosomes 1D, 2D, 4A, 4B, 5A, 5D, 6A, and 6B; with consistent QTL (that are identified in all the years) on chromosome 4A in DH population-1, and on chromosomes 2D and 6B in DH population-2. The consistent QTL explained 17.2%, 32.9%, and 12.3% of the phenotypic variances, respectively. Genic scan of the chromosome 2D-QTL showed that the wheat gene TaAP2-D, an ortholog of Cleistogamy1 which promotes AE via swelling of the lodicules in barley, lies within the QTL region. A diagnostic marker was developed for TaAP2-D that showed co-segregation with the AE phenotype. This study shows the use of gene bank diversity reservoir to find alleles which are otherwise difficult to detect in elite populations. The identification of large-effect consistent QTL for AE is expected to help form efficient male parental lines suitable for hybrid wheat seed production and serve as a source for map-based cloning.




Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Aukerman MJ, Sakai H (2003) Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15:2730–2741
Bailey TB, Qualset CO, Cox DF (1980) Predicting heterosis in wheat. Crop Sci 20:339–342
Barbosa-Neto JF, Sorrells ME, Cisar G (1996) Prediction of heterosis in wheat using coefficient of parentage and RFLP-based estimates of genetic relationship. Genome 39:1142–1149
Beri S, Anand S (1971) Factors affecting pollen shedding capacity in wheat. Euphytica 20:327–332
Bernardo R (2008) Molecular markers and selection for complex traits in plants: learning from the last 20 years. Crop Sci 48:1649–1664
Bernardo R (2010) Breeding for quantitative traits in plants, 2nd edn. Stemma Press, Woodbury
Boeven PH, Longin CFH, Leiser WL, Kollers S, Ebmeyer E, Würschum T (2016) Genetic architecture of male floral traits required for hybrid wheat breeding. Theor Appl Genet 129:2343–2357
Bruns R, Peterson CJ (1998) Yield and stability factors associated with hybrid wheat. Euphytica 100:1–5
Buerstmayr M, Buerstmayr H (2015) Comparative mapping of quantitative trait loci for Fusarium head blight resistance and anther retention in the winter wheat population Capo × Arina. Theor Appl Genet 128:1519–1530
Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138:963–971
Consortium IWGS (2018) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361:eaar7191
Dafni A, Firmage D (2000) Pollen viability and longevity: practical, ecological and evolutionary implications. Plant Syst Evol 222:113–132
De Vries AP (1971) Flowering biology of wheat, particularly in view of hybrid seed production - a review. Euphytica 20:152–170
De Vries AP (1972) Some aspects of cross-pollination in wheat (Triticum aestivum L.) 1. Pollen concentration in the field as influenced by variety, diurnal pattern, weather conditions and level as compared to the height of the pollen donor. Euphytica 21:185–203
De Vries AP (1973) Some aspects of cross-pollination in wheat (Triticum aestivum L.) 2. Anther extrusion and ear and plant flowering pattern and duration. Euphytica 22:445–456
De Vries AP (1974) Some aspects of cross-pollination in wheat (Triticum aestivum L.). 4. Seed set on male sterile plants as influenced by distance from the pollen source, pollinator: male sterile ratio and width of the male sterile strip. Euphytica 23:601–622
D’Souza L (1970) Studies on the suitability of wheat as pollen donor for cross pollination, compared with rye, Triticale and Secalotricum. Zeitschrift fur Pflanzenzüchtung 63:246–269
Graham S, Browne RA (2009) Anther extrusion and Fusarium head blight resistance in European wheat. J Phytopathol 157:580–582
He X, Singh PK, Dreisigacker S, Singh S, Lillemo M, Duveiller E (2016) Dwarfing genes Rht-B1b and Rht-D1b are associated with both type I FHB susceptibility and low anther extrusion in two bread wheat populations. PLoS ONE 11:e0162499
Irish VF, Sussex IM (1990) Function of the apetala-1 gene during Arabidopsis floral development. Plant Cell 2:741–753
Jiang Y, Schmidt RH, Zhao Y, Reif JC (2017) A quantitative genetic framework highlights the role of epistatic effects for grain-yield heterosis in bread wheat. Nat Genet 49:1741
Jofuku KD, den Boer BG, Van Montagu M, Okamuro JK (1994) Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6:1211–1225
Koekemoer FP, Van Eeden E, Bonjean AP (2011) An overview of hybrid wheat production in South Africa and review of current worldwide wheat hybrid developments. World Wheat Book A Hist Wheat Breed 2:907–950
Langer SM, Longin CFH, Würschum T (2014) Phenotypic evaluation of floral and flowering traits with relevance for hybrid breeding in wheat (Triticum aestivum L.). Plant Breed 133:433–441
Li H (2018) Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34:3094–3100
Lord EM (1981) Cleistogamy: a tool for the study of floral morphogenesis, function and evolution. Bot Rev 47:421–449
Lu QX, Lillemo M, Skinnes H, He XY, Shi JR, Ji F, Dong YH, Bjørnstad Å (2013) Anther extrusion and plant height are associated with Type I resistance to Fusarium head blight in bread wheat line ‘Shanghai-3/Catbird’. Theor Appl Genet 126:317–334
Lynch M, Walsh B (1998) Genetics and analysis of quantitative traits. Sinauer Associates Inc, Sunderland
Mühleisen J, Piepho HP, Maurer HP, Longin CFH, Reif JC (2014) Yield stability of hybrids versus lines in wheat, barley, and triticale. Theor Appl Genet 127:309–316
Muqaddasi QH, Lohwasser U, Nagel M, Börner A, Pillen K, Röder MS (2016) Genome-wide association mapping of anther extrusion in hexaploid spring wheat. PLoS ONE 11:e0155494
Muqaddasi QH, Brassac J, Börner A, Pillen K, Röder MS (2017a) Genetic architecture of anther extrusion in spring and winter wheat. Front Plant Sci 8:754
Muqaddasi QH, Pillen K, Plieske J, Ganal MW, Röder MS (2017b) Genetic and physical mapping of anther extrusion in elite European winter wheat. PLoS ONE 12:e0187744
Muqaddasi QH, Reif JC, Li Z, Basnet BR, Dreisigacker S, Röder MS (2017c) Genome-wide association mapping and genome-wide prediction of anther extrusion in CIMMYT spring wheat. Euphytica 213:73
Myles S, Peiffer J, Brown PJ, Ersoz ES, Zhang Z, Costich DE, Buckler ES (2009) Association mapping: critical considerations shift from genotyping to experimental design. Plant Cell 21:2194–2202
Nair SK, Wang N, Turuspekov Y, Pourkheirandish M, Sinsuwongwat S, Chen G, Sameri M, Tagiri A, Honda I, Watanabe Y, Kanamori H, Wicker T, Stein N, Nagamura Y, Matsumoto T, Komatsuda T (2010) Cleistogamous flowering in barley arises from the suppression of microRNA-guided HvAP2 mRNA cleavage. Proc Natl Acad Sci USA 107:490–495
Nettevich ED (1968) The problem of utilizing heterosis of wheat (Triticum aestivum). Euphytica 17:54–62
Ning S, Wang N, Sakuma S, Pourkheirandish M, Koba T, Komatsuda T (2013a) Variation in the wheat AP2 homoeologs, the genes underlying lodicule development. Breed Sci 63:255–266
Ning S, Wang N, Sakuma S, Pourkheirandish M, Wu J, Matsumoto T, Koba T, Komatsuda T (2013b) Structure, transcription and post-transcriptional regulation of the bread wheat orthologs of the barley cleistogamy gene Cly1. Theor Appl Genet 126:1273–1283
Okamuro JK, Caster B, Villarroel R, Van Montagu M, Jofuku KD (1997) The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc Natl Acad Sci 94:7076–7081
Pickett AA (1993) Hybrid wheat—results and problems. Fortschritte der Pflanzenzüchtung. Paul Parey Scientific Publishers, Berlin
Pickett AA, Galwey NW (1997) A further evaluation of hybrid wheat. Plant Var Seeds 10:15–32
Röder MS, Korzun V, Wendehake K, Plaschke J, Tixier M-H, Leroy P, Ganal MW (1998) A microsatellite map of wheat. Genetics 149:2007–2023
Shull GH (1948) What is” heterosis”? Genetics 33:439–446
Singh SK, Arun B, Joshi AK (2007) Comparative evaluation of exotic and adapted germplasm of spring wheat for floral characteristics in the Indo-Gangetic Plains of northern India. Plant Breed 126:559–564
Singh S, Chatrath R, Mishra B (2010) Perspective of hybrid wheat research: a review. Indian J Agric Sci 80:1013–1027
Skinnes H, Semagn K, Tarkegne Y, Maroy AG, Bjørnstad Å (2010) The inheritance of anther extrusion in hexaploid wheat and its relationship to Fusarium head blight resistance and deoxynivalenol content. Plant Breed 129:149–155
Tanksley SD, McCouch SR (1997) Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277:1063–1066
Uphof JCT (1938) Cleistogamic Flowers. Bot Rev 4:21–49
Van Ooijen J (2006) JoinMap® 4, Software for the calculation of genetic linkage maps in experimental populations. Kyazma BV, Wageningen, p 33
Walton PD (1971) Heterosis in spring wheat. Crop Sci 11:422–424
Wang S, Basten CJ, Zeng Z-B (2012) Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC. https://brcwebportal.cos.ncsu.edu/qtlcart/
Wang SC, Wong DB, Forrest K, Allen A, Chao SM, Huang BE, Maccaferri M, Salvi S, Milner SG, Cattivelli L, Mastrangelo AM, Whan A, Stephen S, Barker G, Wieseke R, Plieske J, Lillemo M, Mather D, Appels R, Dolferus R, Brown-Guedira G, Korol A, Akhunova AR, Feuillet C, Salse J, Morgante M, Pozniak C, Luo MC, Dvorak J, Morell M, Dubcovsky J, Ganal M, Tuberosa R, Lawley C, Mikoulitch I, Cavanagh C, Edwards KJ, Hayden M, Akhunov E, Characterization of polyploid wheat genomic diversity using a high-density 90 000 single nucleotide polymorphism array (2014) Characterization of polyploid wheat genomic diversity using a high-density 90 000 single nucleotide polymorphism array. Plant Biotechnol J 12:787–796
Wilson JA (1968) Problems in hybrid wheat breeding. Euphytica 17:13
Wilson J, Ross W (1962) Cross breeding in wheat, Triticum aestivum L. II. Hybrid seed set on a cytoplasmic male-sterile winter wheat composite subjected to cross-pollination. Crop Sci 2:415–417
Zhang X, Xiao Y, Zhang Y, Xia X, Dubcovsky J, He Z (2008) Allelic variation at the vernalization genes Vrn-A1, Vrn-B1, Vrn-D1, and Vrn-B3 in Chinese wheat cultivars and their association with growth habit. Crop Sci 48:458–470
Zhou Y, Lu D, Li C, Luo J, Zhu B-F, Zhu J, Shangguan Y, Wang Z, Sang T, Zhou B, Han B (2012) Genetic control of seed shattering in rice by the APETALA2 transcription factor SHATTERING ABORTION1. Plant Cell 24:1034–1048
Acknowledgements
This study was funded by Grant4Traits, a project by Bayer AG, Division Crop Science. We are grateful to Ellen Weiß, Anette Heber, Ute Ostermann, and Sonja Allner for help in data collection. We thank Ravi Koppolu and Martin Mascher for helpful discussions. The authors gratefully acknowledge the handling editor and three anonymous reviewers whose comments helped to improve this manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standards
On behalf of all co-authors, the corresponding author states that the work described is original, previously unpublished research. All the authors listed have approved the manuscript.
Additional information
Communicated by Steven S. Xu.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Muqaddasi, Q.H., Jayakodi, M., Börner, A. et al. Identification of consistent QTL with large effect on anther extrusion in doubled haploid populations developed from spring wheat accessions in German Federal ex situ Genebank. Theor Appl Genet 132, 3035–3045 (2019). https://doi.org/10.1007/s00122-019-03404-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-019-03404-2