Abstract
Key message
Using next-generation DNA sequencing, it was possible to clarify the genetic relationships of Avena species and deduce the likely pathway from which hexaploid oat was formed by sequential polyploidization events.
Abstract
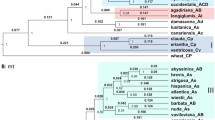
A sequence-based diversity study was conducted on a representative sample of accessions from species in the genus Avena using genotyping-by-sequencing technology. The results show that all Avena taxa can be assigned to one of four major genetic clusters: Cluster 1 = all hexaploids including cultivated oat, Cluster 2 = AC genome tetraploids, Cluster 3 = C genome diploids, Cluster 4 = A genome diploid and tetraploids. No evidence was found for the existence of discrete B or D genomes. Through a series of experiments involving the creation of in silico polyploids, it was possible to deduce that hexaploid oat likely formed by the fusion of an ancestral diploid species from Cluster 3 (A. clauda, A. eriantha) with an ancestral diploid species from Cluster 4D (A. longiglumis, A. canariensis, A. wiestii) to create the ancestral tetraploid from Cluster 2 (A. magna, A. murphyi, A. insularis). Subsequently, that ancestral tetraploid fused again with another ancestral diploid from Cluster 4D to create hexaploid oat. Based on the geographic distribution of these species, it is hypothesized that both the tetraploidization and hexaploidization events may have occurred in the region of northwest Africa, followed by radiation of hexaploid oat to its current worldwide distribution. The results from this study shed light not only on the origins of this important grain crop, but also have implications for germplasm collection and utilization in oat breeding.






Similar content being viewed by others
References
Chen Q, Armstrong K (1994) Genomic in situ hybridization in Avena sativa. Genome 37:607–612
Cooper JK, Ibrahim AMH, Rudd J, Hays D, Malla S, Baker J (2013) Increasing hard winter wheat yield potential via synthetic hexaploid wheat: II. heritability and combining ability of yield and its components. Crop Sci 53:67–73
Elshire RJ, Glaubitz JC, Sun Q, Poland J, Kawamoto K, Buckler ES, Mitchell SE (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6(5):e-19379
Fominaya A, Vega C, Ferrer E (1988) C-banding and nucleolar activity of tetraploid Avena species. Genome 30:633–638
Fu Y, Williams D (2008) AFLP variation in 25 Avena species. Theor Appl Genet 117:333–342
Girke A, Schierholt A, Becker HC (2012) Extending the rapeseed gene pool with resynthesized Brassica napus II: heterosis. Theor Appl Genet 124:1017–10216
Jellen EN, Bill BS, Cox TS (1994) Genomic in situ hybridization differentiates between A/D and C genomes chromatin and detects intergenomic translocations in polyploid oat species. Genome 37:613–618
Jesske T, Olberg B, Schierholt A, Becker HC (2013) Resynthesized lines from domesticated and wild Brassica taxa and their hybrids with B. napus L.: genetic diversity and hybrid yield. Theor Appl Genet 126:1053–1065
Kim KY et al (2008) Rapid chromosome evolution in recently formed polyploids in Tragopogon (Asteraceae). PLoS One 3:e3353
Ladizinsky GA (1998) A new species of Avena from Sicily, possibly the tetraploid progenitor of hexaploid oat. Gene Resour Crop Evol 45:263–269
Ladizinsky G (1999) Cytogenetic relationships between Avena insularis (2n = 28) and A. strigosa (2n = 14) and A. murphyi (2n = 28). Gene Resour Crop Evol 46:501–504
Ladizinsky G (2000) A synthetic hexaploid (2n = 42) oat from the cross of Avena strigosa (2n = 14) and domesticated A. magna (2n = 28). Euphytica 116:231–235
Ladizinsky G (2012) Studies in oat evolution: a man’s life with Avena. Springer, New York
Ladizinsky G, Zohary D (1968) Genetic relationships between diploids and tetraploids iin Series Eubarbatae of Avena. Can J Genet Cytol 10:68–81
Leggett JM (1992) Classification and speciation in Avena. In: Marshall HG, Sorrells ME (eds) Oat science and technology. Agronomy monograph 33. American Society of Agronomy (ASA) and Crop Science Society of America (CSSA), Madison, pp 29–52
Leggett JM, Markhand SM (1995) The genomic identification of some monosomics of Avena sativa L cv. Sun II using GISH. Sun II using GISH. Genome 38:747–751
Leggett JM, Thomas H (1995) Oat evolution and cytogenetics. In: Welch W (ed) The oat crop. Production and utilization. Chapman & Hall, London, pp 121–149
Leggett JM, Thomas HM, Meredith MR, Humphreys MW, Morgan WG, Thomas H, King IP (1994) Intergenomic Translocation and the genomic composition of Avena maroccana Gdgr. revealed by FISH. Chromosome Res 2:163–164
Li C, Rossnagel BG, Scoles GJ (2000) Tracing the phylogeny of the hexaploid oat Avena sativa with satellite DNAs. Crop Sci 40:1755–1763
Linares C, Gonzalez J, Ferrer E, Fominaya A (1996) The use of double in situ hybridization to physically map the positions of 5S rDNA genes in relation to the chromosomal location of 18S-5.8S-26S rDNA and a C genome specific DNA sequence in the genus Avena. Genome 39:535–542
Linares C, Ferrer E, Fominaya A (1998) Discrimination of the closely related A and D genomes of the hexaploid oat Avena sativa L. Proc Natl Acad Sci USA 95:12450–12455
Lopes MS, Reynolds MP (2011) Drought adaptive traits and wide adaptation in elite lines derived from resynthesized hexaploid wheat. Crop Sci 51:1617–1626
Loskutov IG (2008) On evolutionary pathways of Avena species. Genet Resour Crop Evol 55:211–220
Loskutov IG, Rines HW (2011) Avena. In: Kole C (ed) Wild crop relatives: genomic and breeding resources: cereals. Springer, New York, pp 109–184
Lu F, Lipka A, Glaubitz J, Elshire R, Cherney JH, Casler MD, Buckler ES (2013) Switchgrass genomic diversity, ploidy, and evolution: novel insights from a network-based SNP discovery protocol. PLoS Genet 9:e1003215
Peng Y, Baum BR, Ren C, Jiang Q, Chen G, Zheng Y, Wei Y (2010) The evolution pattern of rDNA ITS in Avena and phylogenetic relationships of the Avena species (Poaceae:Avenae). Hereditas 147:183–204
Pires JC et al (2004) Flowering time divergence and genomic rearrangements in resynthesized Brassica polyploids (Brassicaceae). Biol J Linn Soc Lond 82:675–688
Pontes O et al (2004) Chromosomal locus rearrangements are a rapid response to formation of the allotetraploid Arabidopsis suecica genome. Proc Natl Acad Sci USA 101:18240–18245
Rajhathy T (1961) Chromosomal differentiation and speciation in diploid Avena. Can J Genet Cytol 3:372–377
Rajhathy T, Morrison JW (1959) Chromosome morphology in the genus Avena. Can J Bot 37:331–337
Rajhathy T, Thomas H (1974) Cytogenetics of oat (Avena L.). Misc Publ Genet Soc Can 2:1–90
Sadasivaiah RS, Rajhathy T (1968) Genome relationships in tetraploid Avena. Can J Genet Cytol 10:655–669
Sanchez de lal Hoz PS, Fominaya A (1989) Studies of isozymes in oat species. Theor Appl Genet 77:735–741
Sanz MJ, Jellen EN, Loarce Y, Irigoyen ML, Ferrer E, Fominaya A (2010) A new chromosome nomenclature system for oat (Avena sativa L. and A. byzantine C. Koch) based on FISH analysis of monosomic lines. Theor Appl Genet 121:1541–1552
Shelukhina OY, Badaeva ED, Loskutov IG, Pukhal’sky VA (2007) A comparative cytogenetic study of the tetraploid oat species with the A and C genomes: Avena insularis, A. magna, A. murphyi. Genetika 43:747–761
Thomas H (1992) Cytogenetics of Avena. In: Marshall HG, Sorrells ME (eds) Oat science and technology., Monograph 33, agronomy seriesASA and CSSA, Madison, pp 473–507
Xiong Z, Gaeta RT, Pires JC (2011) Homoeologous shuffling and chromosome compensation maintain genome balance in resynthesized allopolyploid Brassica napus. Proc Natl Acad Sci 108:7908–7913
Acknowledgments
This research was funded by PepsiCo Global R&D. The views expressed in this article are those of the authors and do not necessarily reflect the position or policy of PepsiCo Inc. Thanks to Don Smith for assistance in curating and analyzing the DNA sequence data and to Peter Eckstein and Mark Colin for plant tissue sampling. We also thank GRIN USDA, GRIN Canada, IPK Gatersleben and John Innes Centre (GRU) for providing the seed stocks used in this study. The BRC has acknowledgement guideline http://www.biotech.cornell.edu/brc/brc/services/terms-and-policies#Acknowledgement.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by M. E. Sorrells.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chew, P., Meade, K., Hayes, A. et al. A study on the genetic relationships of Avena taxa and the origins of hexaploid oat. Theor Appl Genet 129, 1405–1415 (2016). https://doi.org/10.1007/s00122-016-2712-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-016-2712-4