Abstract
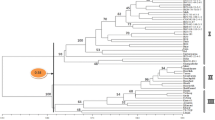
Current molecular characterization of ex situ plant germplasm has placed more emphasis on cultivated gene pools and less on exotic gene pools representing wild relative species. This study attempted to characterize a selected set of germplasm accessions representing various Avena species with the hope to establish a reference set of exotic oat germplasm for oat breeding and research. The amplified fragment length polymorphism (AFLP) technique was applied to screen 163 accessions of 25 Avena species with diverse geographic origins. For each accession, 413 AFLP polymorphic bands detected by five AFLP primer pairs were scored. The frequencies of polymorphic bands ranged from 0.006 to 0.994 and averaged 0.468. Analysis of molecular variance revealed 59.5% of the total AFLP variation resided among 25 oat species, 45.9% among six assessed sections of the genus, 36.1% among three existing ploidy levels, and 50.8% among eight defined genome types. All the species were clustered together according to their ploidy levels. The C genome diploids appeared to be the most distinct, followed by the Ac genome diploid A. canariensis. The Ac genome seemed to be the oldest in all the A genomes, followed by the As, Al and Ad genomes. The AC genome tetraploids were more related to the ACD genome hexaploids than the AB genome tetraploids. Analysis of AFLP similarity suggested that the AC genome tetraploid A. maroccana was likely derived from the Cp genome diploid A. eriantha and the As genome diploid A. wiestii, and might be the progenitor of the ACD genome hexaploids. These AFLP patterns are significant for our understanding of the evolutionary pathways of Avena species and genomes, for establishing reference sets of exotic oat germplasm, and for exploring new exotic sources of genes for oat improvement.




Similar content being viewed by others
References
Alicchio R, Aranci L, Conte L (1995) Restriction fragment length polymorphism based phylogenetic analysis of Avena L. Genome 38:1279–1284
Althoff DM, Gitzendanner MA, Segraves KA (2007) The utility of amplified fragment length polymorphism in phylogenetics: a comparison of homology within and between genomes. Syst Biol 56:477–484
Baum BR (1977) Oats: wild and cultivated, a monograph of the Genus Avena L. (Poaceae). Thorn Press Limited, Ottawa
Baum BR, Fedak G (1985a) Avena altantica, a new diploid species of the oat genus from Morocco. Can J Bot 63:1057–1060
Baum BR, Fedak G (1985b) A new tetraploid species of Avena discovered in Morocco. Can J Bot 63:1379–1385
Chen Q, Armstrong K (1994) Genomic in situ hybridization in Avena sativa. Genome 37:607–612
Dice LR (1945) Measures of the amount of ecologic association between species. Ecology 26:297–302
Diederichsen A, Timmermans E, Williams DJ, Richards KW (2001) Holding of Avena germplasm at plant gene resources of Canada and status of the collection. Oat Newsl 47:35–42
Drossou A, Katsiotis A, Leggett JM, Loukas M, Tsakas S (2004) Genome and species relationships in genus Avena based on RAPD and AFLP molecular markers. Theor Appl Genet 109:48–54
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.01: an integrated software package for population genetics data analysis. Evol Bioinform Online 1:47–50
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Frey KJ (1991) Genetic resources of oats. In: Shands HL, Wiesner L (eds) Use of plant introduction in cultivar development. Crop Science Society of America Special Publication No. 17, Madison, WI, USA, pp 15–24
Fridman E, Carrari F, Liu Y-S, Fernie AR, Zamir D (2004) Zooming in on a quantitative trait for tomato yield using interspecific introgressions. Science 305:1786–1789
Fu YB, Kibite S, Richards KW (2004) Amplified fragment length polymorphism analysis of 96 Canadian oat cultivars released between 1886 and 2001. Can J Plant Sci 84:23–30
Fu YB, Peterson GW, Williams D, Richards KW, Fetch JM (2005) Patterns of AFLP variation in a core subset of cultivated hexaploid oat germplasm. Theor Appl Genet 111:530–539
Fu YB, Chong J, Fetch T, Wang ML (2007) Microsatellite variation in germplasm accessions of the wild oat Avena sterilis L. Theor Appl Genet 114:1029–1038
Harder DE, Chong J, Brown PD, Sebesta J, Fox S (1992) Wild oat as a source of disease resistance: history, utilization, and prospects. In: Proceedings of the 4th International Oat Conference, vol II. Wild Oats in World Agriculture, Adelaide, Australia, pp 71–81
Harlan JR, De Wet JMJ (1971) Toward a rational classification of cultivated plants. Taxon 20:509–517
Hawkes JG (1990) The potato: evolution biodiversity and genetic resources. Smithsonian Institution Press, Washington, DC
Hodkinson TR, Renvoize SA, Chonghaile GN, Stapleton CMA, Chase MW (2000) A comparison of ITS nuclear rDNA sequence data and AFLP markers for phylogenetic studies in Phyllostachys (Bambusoideae, Poaceae). J Plant Res 113:259–269
Irigoyen ML, Loarce Y, Linares C, Ferrer E (2001) Discrimination of the closely related A and B genomes in AABB tetraploid species of Avena. Theor Appl Genet 103:1160–1166
Jellen EN, Gill BS, Cox TS (1994) Genomic in situ hybridization differentiates between A/D- and C-genome chromatin and detects intergenomic translocations in polyploidy oat species (genus Avena). Genome 37:613–618
Jellen EN, Leggett JM (2006) Cytogenetic manipulation in oat improvement. In: Singh RJ, Jauhar PP (eds) Genetic resources, chromosome engineering, and crop improvement, cereals, vol 2. Taylor and Francis, Boca Raton, pp 199–231
Karp A (2002) The new genetic era: will it help us in managing genetic diversity? In: Engels JMM, Rao VR, Brown AHD, Jackson MT (eds) Managing plant genetic diversity. International Plant Genetic Resources Institute, Rome, pp 43–56
Katsiotis A, Hagidimitriou M, Heslop-Harrison JS (1997) The close relationship between the A and B genomes in Avena L. (Poaceae) determined by molecular cytogenetic analysis of total genomic, tandemly and dispersed repetitive DNA sequences. Ann Bot 79:103–109
Koopman WJM (2005) Phylogenetic signal in AFLP data sets. Syst Biol 54:197–217
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Ladizinsky G (1971) Biological flora of Israel II. Avena L. Israel J Bot 20:133–151
Ladizinsky G (1988) Biological species and wild genetic resources in Avena. In: Mattsson B, Lyhagen R (eds) Proceedings of the 3rd International Oat Conference, Svalöf AB, Sweden, pp 76–86
Ladizinsky G (1998) A new species of Avena from Sicily, possibly the tetraploid progenitor of hexaploid oats. Genet Resour Crop Evol 45:263–269
Ladizinsky G (1999) Cytogenetic relationships between Avena insularis (2n = 28) and both A. strigosa (2n = 14) and A. murphyi (2n = 28). Genet Resour Crop Evol 46:501–504
Leggett JM (1992) Classification and speciation in Avena. In: Marshall HG, Sorrells ME (eds) Oat science and technology. Monograph 33, Agronomy series. ASA and CSSA, Madison, pp 29–52
Leggett JM (1996) Using and conserving Avena genetic resources. In: Scoles GJ, Rossnagel BG (eds) Proceedings of the Vth international oat conference and VIIth international barley genetic symposium, University of Saskatchewan, Saskatoon, pp 128–132
Leggett JM, Markhand GS (1995) The genomic structure of Avena revealed by GISH. In: Brandham PE, Bennett MD (eds) Kew chromosome conference IV. HMSO, UK, pp 133–139
Leggett JM, Thomas H (1995) Oat evolution and cytogenetics. In: Welch W (ed) The oat crop: production and utilization. Chapman & Hall, London, pp 121–149
Li CD, Rossnagel BG, Scoles GJ (2000) Tracing the phylogeny of the hexaploid oat Avena sativa with satellite DNAs. Crop Sci 40:1755–1763
Linares C, Ferrer E, Fominaya A (1998) Discrimination of the closely related A and D genomes of the hexaploid oat Avena sativa L. Proc Natl Acad Sci USA 95:12450–12455
Loskutov IG (2008) On evolutionary pathways of Avena species. Genet Resour Crop Evol 55:211–220
Loskutov IG, Perchuk IN (2000) Evaluation of interspecific diversity in Avena genus by RAPD analysis. Oat Newsl 46:38–41
Malzew AI (1930) Wild and cultivated oats Section Eu Avena Griseb. Bull Appl Bot Genet Plant Breed Suppl 38th, Leningrad, 522 pp
Mechanda SM, Baum BR, Johnson DA, Arnason JT (2004) Sequence assessment of comigrating AFLPTM bands in Echinacea–implications for comparative studies. Genome 47:15–25
Nocelli E, Giovannini T, Bioni M, Alicchio R (1999) RFLP- and RAPD-based genetic relationships of several diploid species of Avena with the A genome. Genome 42:950–959
Rajhathy T, Thomas H (1974) Cytogenetics of oats (Avena L.). Genetic Soc Can Misc Publ 2, Ottawa
de la Hoz Sanchez P, Fominaya A (1989) Studies of isozymes in oat species. Theor Appl Genet 77:735–741
Institute Inc. SAS (2004) The SAS system for windows V8.02. SAS Institute Incorporated, Cary
Swofford DL (1998) PAUP*. Phylogenetic analysis using parsimony (*and other methods). version 4. Sinauer Associates, Sunderland
Tanksley SD, McCouch SR (1997) Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277:1063–1066
Thomas H (1992) Cytogenetics of Avena. In: Marshall HG, Sorrells ME (eds) Oat science and technology. Monograph 33, Agronomy series. ASA and CSSA, Madison, pp 473–507
Thomas H (1995) Oats. In: Smartt J, Simmonds NW (eds) Evolution of crop plants. Longman Group, New York, pp 132–140
Vos P, Hogers R, Bleeker M, Reijans M, van De Lee T, Hornes M, Frijters A, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Wesenberg DM, Briggle LW, Smith DH (1992) Germplasm collection, preservation and utilization. In: Marshall HG, Sorrells ME (eds) Oat science and technology. Monograph 33, Agronomy series. ASA and CSSA, Madison, pp 793–818
Acknowledgments
Authors would like to thank Dallas Kessler, Gregory Peterson and Marcie Heggie for their technical assistance in sampling, planting, and genotyping the germplasm; and Richard Gugel, Jie Qiu and two anonymous reviewers for their helpful comments on the early versions of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Morgante.
Electronic supplementary material
Below is the link to the electronic supplementary material.
122_2008_778_MOESM1_ESM.doc
Supplementary Table S1 List of 163 Avena accessions assayed with information on ploidy, genome type, origin, Canadian National (CN) accession number and label used in Fig. 2 for this study (doc 47 KB)
Rights and permissions
About this article
Cite this article
Fu, YB., Williams, D.J. AFLP variation in 25 Avena species. Theor Appl Genet 117, 333–342 (2008). https://doi.org/10.1007/s00122-008-0778-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-008-0778-3