Abstract
Key message
A major quantitative trait locus (QTL) for Fusarium oxysporum Fr. f. sp. niveum race 1 resistance was identified by employing a “selective genotyping” approach together with genotyping-by-sequencing technology to identify QTLs and single nucleotide polymorphisms associated with the resistance among closely related watermelon genotypes.
Abstract
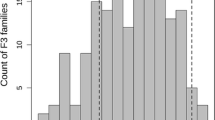
Fusarium wilt is a major disease of watermelon caused by the soil-borne fungus Fusarium oxysporum Schlechtend.:Fr. f. sp. niveum (E.F. Sm.) W.C. Snyder & H.N. Hans (Fon). In this study, a genetic population of 168 F3 families (24 plants in each family) exhibited continuous distribution for Fon race 1 response. Using a “selective genotyping” approach, DNA was isolated from 91 F2 plants whose F3 progeny exhibited the highest resistance (30 F2 plants) versus highest susceptibility (32 F2 plants), or moderate resistance to Fon race 1 (29 F2 plants). Genotyping-by-sequencing (GBS) technology was used on these 91 selected F2 samples to produce 266 single nucleotide polymorphism (SNP) markers, representing the 11 chromosomes of watermelon. A major quantitative trait locus (QTL) associated with resistance to Fon race 1 was identified with a peak logarithm of odds (LOD) of 33.31 and 1-LOD confidence interval from 2.3 to 8.4 cM on chromosome 1 of the watermelon genetic map. This QTL was designated “Fo-1.1” and is positioned in a genomic region where several putative pathogenesis-related or putative disease-resistant gene sequences were identified. Additional independent, but minor QTLs were identified on chromosome 1 (LOD 4.16), chromosome 3 (LOD 4.36), chromosome 4 (LOD 4.52), chromosome 9 (LOD 6.8), and chromosome 10 (LOD 5.03 and 4.26). Following the identification of a major QTL for resistance using the “selective genotyping” approach, all 168 plants of the F 2 population were genotyped using the SNP nearest the peak LOD, confirming the association of this SNP marker with Fon race 1 resistance. The results in this study should be useful for further elucidating the mechanism of resistance to Fusarium wilt and in the development of molecular markers for use in breeding programs of watermelon.



Similar content being viewed by others
References
Akram A, Ongena M, Duby F, Dommes J, Thonart P (2008) Systemic resistance and lipoxygenase-related defence response induced in tomato by Pseudomonas putida strain BTP1. BMC Plant Biol 8:113. doi:10.1186/1471-2229-8-113
Arumuganathan K, Earle ED (1991) Nuclear DNA content of some important plant species. Plant Mol Biol Rep 9:208–218
Bruton BD, Damicone JP (1999) Fusarium wilt of watermelon: impact of race 2 of Fusarium oxysporum f. sp. niveum on watermelon production in Texas and Oklahoma. Subtrop Plant Sci 51:4–9
Bruton BD, Patterson CL, Martyn RD (1988) Fusarium wilt (Fusarium oxysporum f. sp. niveum race 2) of watermelons in Oklahoma. Plant Dis 72:734
Dane F, Hawkins LK, Norton JD (1998) New resistance to Race 2 of Fusarium oxysporum F. sp. niveum in watermelon. Cucurbit Genet Coop Rep 21:27–29
Darvasi A, Soller M (1992) Selective genotyping for determination of linkage between a marker locus and a quantitative trait locus. Theor Appl Genet 85:353–359
Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 6:e19379
Esposito RG, Fletcher AM (1961) The relationship of pteridine biosynthesis to the action of copper 8-hdroxyquinolate on fungal spores. Arch Biochem Biophys 93:369–376
Goff KE, Ramonell KM (2007) The role and regulation of receptor-like kinases in plant defense. Gene Regul and Syst Biol 1:167–175
Guo S, Zhang J, Sun H, Salse J, Lucas WJ, Zheng Y, Mao L, Ren Y, Wang Z, Min J, Zhang H, Murat F, Ham B, Zhang Z, Gao S, Huang M, Xu Y, Zhong S, Zhao H, He H, Zhang Y, Zhang Z, Huang S, Tan T, Pang E, Lin K, Hu Q, Kuang H, Liu J, Kou Q, Hou W, Zou X, Jiang J, Gong G, Klee K, Schoof H, Huang Y, Ni P, Wang B, Wang J, Wu K, Xia Y, Zhao X, Zheng Z, Xing M, Liang X, Huang B, Lv T, Wang J, Yi H, Li R, Wu M, Levi A, Zhang X, Giovannoni JJ, Wang J, Li Y, Fei Z, Xu Y (2013) Genome sequencing of watermelon (Citrullus lanatus): insights into evolution, domestication and crop improvement. Nat Genet 45:51–58
Gusmini G, Song R, Wehner TC (2005) New sources of resistance to Gummy Stem Blight in watermelon. Crop Sci 45:582–588
Harris KR, Wechter WP, Levi A (2009) Isolation, sequence analysis, and linkage mapping of NBS–LRR disease resistance gene analogs in watermelon. J Am Soc Hortic Sci 134:649–657
Harris-Shultz KR, Wechter WP, Levi A (2010) Linkage mapping of NBS–LRR disease resistance gene analogs in watermelon. HortScience 45:510
Hou M, Xu W, Bai H, Liu Y, Li L, Liu L, Liu B, Liu G (2012) Characteristic expression of rice pathogenesis-related proteins in rice leaves during interactions with Xanthomonas oryzae pv. oryzae. Plant Cell Rep 31:895–904
Keinath AP, Hassell RL, Everts KL, Zhou XG. (2010) Cover crops of hybrid common vetch reduce Fusarium wilt of seedless watermelon in the eastern United States. Online. Plant Health Prog. 20 Feb 2010. http://www.plantmanagementnetwork.org/sub/php/research/2010/vetch
Lander E, Botstein D (1989) Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121:185–199
Lebowitz RJ, Soller M, Beckmann JS (1987) Trait-based analyses for the detection of linkage between marker loci and quantitative trait loci in crosses between inbred lines. Theor Appl Genet 73:556–562
Levi A, Thomas CE, Wehner TC, Zhang X (2001) Low genetic diversity indicates the need to broaden the genetic base of cultivated watermelon. HortScience 36:1096–1101
Levi A., Hernandez A, Thimmapuram J, Donthu R, Wright C, Ali C, Wechter WP, Reddy U, Mikel M (2011) Sequencing the genome of the heirloom watermelon cultivar Charleston Gray. Plant Anim Genome Conf P047
Levi A, Thies J, Wechter WP, Kousik C, Ling K, Harrison H, Simmons A, Reddy UK, Nimmakayala P, Fei Z, Mitchell S, Xu Y, Tadmor K, Katzir N (2012) Exploiting genetic diversity in Citrullus spp. to enhance watermelon cultivars. In: Nabahat S (ed) Xth Eucarpia international meeting on Cucurbitaceae
Levi A, Thies JA, Wechter WP, Harrison HF Jr, Simmons AM, Reddy U, Nimmakayala P, Fei Z (2013) High frequency oligonucleotides—targeting active gene (HFO-TAG) markers reveal wide genetic diversity among Citrullus spp. accessions useful for enhancing disease or pest resistance in watermelon cultivars. Genet Resour Crop Evol 60:427–440
Mackay TFC (2001) Quantitative trait loci in drosophila. Nat Rev Genet 2:11–20
Martyn RD (1985) An aggressive race of Fusarium oxysporum f. sp. niveum new to the United States. Plant Dis 69:1007
Martyn RD (1987) Fusarium oxysporum f. sp. niveum race 2: a highly aggressive race new to the United States. Plant Dis 71:233–236
Martyn RD, Bruton BD (1989) An initial survey of the United States for races of Fusarium oxysporum f. sp. niveum. HortScience 24:696–698
Martyn RD, Netzer D (1991) Resistance to races 0, 1, and 2 of Fusarium wilt of watermelon in Citrullus sp. PI-296341-FR. HortScience 26:429–432
Martyn RD (1996) Fusarium wilt of watermelon. In: Zitter TA, Hopkins DL, Thomas CE (eds) Compendium of cucurbit diseases. APS Press, American Phytopathological Society, St. Paul, pp 13–14
Mazzeo MF, Cacace G, Ferriello F, Puopolo G, Zoina A, Ercolano ME, Siciliano RA (2014) Proteomic investigation of response to FORL infection in tomato roots. Plant Physiol Biochem 74:42–49
Michelmore RW, Paran I, Kesseli RV (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA 88:9828–9832
Netzer D (1976) Physiological races of soil population levels of Fusarium wilt of watermelon. Phytoparasitica 4:131–136
Netzer D, Martyn RD (1989) PI 296341, a source of resistance in watermelon to race 2 of Fusarium oxysporum f. sp. niveum. Plant Dis 73:518
Ren Y, McGregor C, Zhang Y, Gong G, Zhang H, Guo S, Sun H, Cai W, Zhang J, Xu Y (2014) An integrated genetic map based on four mapping populations and quantitative trait loci associated with economically important traits in watermelon ( Citrullus lanatus ). BMC Plant Biol 14:33. doi:10.1186/1471-2229-14-33
Roff DA (2007) A centennial celebration for quantitative genetics. Evolution 61:1017–1032
van Loon LC, Rep M, Pieterse CMJ (2006) Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol 44:135–162
Van Ooijen JW (2009) MAPQTL 6, Software for the mapping of quantitative trait loci in experimental populations of diploid species. Kyazma BV, Wageningen, The Netherlands
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78
Wechter WP, Dean RA, Thomas CT (1998) Development of sequence-specific primers that amplify a 1.5-kb DNA marker for race 1 Fusarium wilt resistance in Cucumis melo L. HortScience 33:291–292
Wechter WP, Kousik CS, McMillan ML, Levi A (2012) Identification of resistance to Fusarium oxysporum f. sp. niveum race 2 in Citrullus lanatus var. citroides plant introductions. HortScience 47:334–338
Yan L, Zhai Q, Wei J, Li S, Wang B, Huang T, Du D, Sun J, Kang L, Li CB, Li C (2013) Role of tomato lipoxygenase D in wound-induced jasmonate biosynthesis and plant immunity to insect herbivores. PLoS Genet 9:e1003422
Zhou XG, Everts KL, Bruton BD (2010) Race 3, a new and highly virulent race of Fusarium oxysporum f. sp. niveum causing Fusarium wilt in watermelon. Plant Dis 94:92–98
Acknowledgments
We thank Sharon Mitchell and the staff at the Institute of Genomic Diversity, Cornell University for performing the genotyping-by-sequencing (GBS) analysis. We thank Zhangjun Fei at the Boyce Thompson Institute for his bioinformatics support. We thank Anthony Keinath and Sandra Pelc for their critical review of this manuscript.
Conflict of interest
This research project was developed through a non-funded cooperative agreement (ARS Number: 58-6659-8-102) between HM.CLAUSE Seed Company and USDA-ARS. The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Michael J. Havey.
Rights and permissions
About this article
Cite this article
Lambel, S., Lanini, B., Vivoda, E. et al. A major QTL associated with Fusarium oxysporum race 1 resistance identified in genetic populations derived from closely related watermelon lines using selective genotyping and genotyping-by-sequencing for SNP discovery. Theor Appl Genet 127, 2105–2115 (2014). https://doi.org/10.1007/s00122-014-2363-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-014-2363-2