Abstract
Little is known about the genetic control of heterosis in the complex polyploid crop species oilseed rape (Brassica napus L.). In this study, two large doubled-haploid (DH) mapping populations and two corresponding sets of backcrossed test hybrids (THs) were analysed in controlled greenhouse experiments and extensive field trials for seedling biomass and yield performance traits, respectively. Genetic maps from the two populations, aligned with the help of common simple sequence repeat markers, were used to localise and compare quantitative trait loci (QTL) related to the expression of heterosis for seedling developmental traits, plant height at flowering, thousand seed mass, seeds per silique, siliques per unit area and seed yield. QTL were mapped using data from the respective DH populations, their corresponding TH populations and from mid-parent heterosis (MPH) data, allowing additive and dominance effects along with digenic epistatic interactions to be estimated. A number of genome regions containing numerous heterosis-related QTL involved in different traits and at different developmental stages were identified at corresponding map positions in the two populations. The co-localisation of per se QTL from the DH population datasets with heterosis-related QTL from the MPH data could indicate regulatory loci that may also contribute to fixed heterosis in the highly duplicated B. napus genome. Given the key role of epistatic interactions in the expression of heterosis in oilseed rape, these QTL hotspots might harbour genes involved in regulation of heterosis (including fixed heterosis) for different traits throughout the plant life cycle, including a significant overall influence on heterosis for seed yield.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Oilseed rape (Brassica napus L; genome AACC, 2n = 38) is the most important source of vegetable oil in Europe and the second most important oilseed crop in the world after soybean. The importance of restored hybrid varieties with improved yield stability and adaptability has increased dramatically during the past decade, and today hybrids have the largest market share in most major oilseed rape and canola growing areas worldwide. However, the yield gain of hybrid varieties compared to homozygous varieties is relatively low compared to classical hybrid crops like maize or rice. In official plot trials in Germany, winter oilseed rape hybrids achieve a mean yield advantage of only around 6% compared to open-pollinated varieties (cumulative data from state variety trials), whereas in practical winter oilseed rape production the 10-year mean yield advantage of hybrids is currently around 11% (data from Kleffmann Group/Norddeutsche Pflanzenzucht H.-G. Lembke KG). A significant improvement in yield performance through more efficient exploitation of heterosis is therefore an important breeding aim.
The genetic basis of heterosis in B. napus is not well understood, however, and few studies have systematically compared the expression and control of heterosis in different genetic backgrounds in this important crop species. Through quantitative trait loci (QTL) mapping of yield-related traits in different populations of backcrossed test hybrids (THs), Quijada et al. (2006) and Udall et al. (2006) revealed a number of common genome regions in which alleles from genetically diverse germplasm contributed significantly to improve yield performance. This suggested that numerous QTL for different yield-related traits in B. napus may overlap at the same or similar positions in different genetic backgrounds. Chen et al. (2007) also identified B. napus chromosome regions harbouring QTL for multiple yield-related traits in a population of B. napus hybrids, and similar QTL hotspots were found by Radoev et al. (2008) to be involved in heterosis for yield-related traits. Interestingly, in all of these four studies, many of the chromosomes containing overlapping yield-related QTL appeared to correspond, with prevalent QTL hotspots being consistently found on chromosomes N01/N11, N03/N13, N05/N15, N07/N16, N12 and N10/N19.
In the model crucifer Arabidopsis thaliana, significant heterosis for numerous biomass-related traits is observed during early seedling developmental stages (Meyer et al. 2004), demonstrating the usefulness of seedlings as a model system to investigate heterosis under more or less controlled conditions. Analysis of biomass and metabolite QTL in Arabidopsis TH populations revealed hotspots of heterotic QTL potentially controlled by regulatory genes (Meyer et al. 2009; Lisec et al. 2009). To our knowledge, no previous study has performed a QTL analysis of heterosis for seedling traits in B. napus, nor attempted to compare per se or heterosis-related QTL for seedling traits with corresponding QTL regions for yield-related traits or seed yield heterosis. In this study, we used two large populations of DH lines, along with corresponding backcross TH populations, for comparative analysis of heterosis-related QTL for seedling and yield traits. The two different DH/TH populations, along with the parental genotypes, were tested independently for different seedling traits in two separate greenhouse trials, then together for yield traits in large-scale field trials at four locations for 2 years. The data generated were used to compare whether per se and heterosis-related QTL for seedling and yield-related traits, at completely different developmental stages, could be detected at common chromosomal positions in different genetic backgrounds and in very different environments.
Materials and methods
Plant materials
The study was performed using two populations of 250 doubled-haploid (DH) lines each, and two corresponding populations of 250 backcrossed THs each. In order to improve map alignment and more easily compare the effects of heterosis-related QTL in different genetic backgrounds, the two DH populations shared the common parent ‘Express 617’, an inbred line derived from the German winter oilseed rape variety ‘Express’. The first DH population (hereinafter referred to as “ExV8-DH”), was generated from a single F1 plant from a cross between ‘Express 617’ and the high erucic acid winter rapeseed inbred line ‘V8’. The adapted breeding line ‘V8’ was derived by recurrent selection from a cross between a high erucic acid winter rapeseed and a resynthesised B. napus that was generated by embryo rescue from an interspecific cross between Brassica oleracea var. botrytis (cauliflower) and Brassica rapa ssp. triloculoris (Yellow Sarson). The second DH population (“ExR53-DH”) was produced from a single F1 plant from a cross between ‘Express 617’ and the resynthesised rapeseed line ‘R53’, an interspecific hybrid between B. oleracea var. acephela (kale) and B. rapa ssp. pekinensis (Chinese cabbage). Due to the diverse origins of ‘V8’ and ‘R53’, the two DH populations were expected to contain considerable allelic diversity despite the common parent ‘Express 617’. Microspore cultivation of DH plants from both populations was performed by Saatenunion Resistenzlabor GmbH, Leopoldshöhe, Germany.
Two corresponding TH populations, referred to hereinafter as “ExV8-TH” and “ExR53-TH”, were generated by crossing the respective DH lines of the ExV8-DH and ExR53-DH populations to the common male-sterile tester ‘MSL-Express’, an isogenic line from ‘Express’ that carries the Male Sterility Lembke (MSL) genic male sterility system. Based on marker analyses, the mother line has up to 96% identity to ‘Express’ (information from Norddeutsche Pflanzenzucht H.-G. Lembke KG), so that the test crosses could be treated as a backcross population. Test crosses were performed in the field in the 2004/2005 growing season.
Because we used isogenic lines of ‘Express’ both as a parent in each of the two DH populations and as the tester for the respective TH populations, the genetic architecture of the THs is identical to a backcross. Any given locus will either be homozygous for ‘Express’ alleles or heterozygous with one ‘Express’ allele and one allele from the other parent of the respective DH population. Because the THs have a maximum of two alleles at each locus, and these alleles can be assigned to the respective parents in the genetic map, it is possible to estimate additive and dominance effects of QTL contributing to heterosis and to calculate digenic epistatic interactions.
Greenhouse trials
The two different populations of DH lines and THs were analysed for seedling biomass traits in independent greenhouse trials at the Universities of Giessen and Göttingen in 2006. In Giessen, the 250 ExV8-DH lines and their corresponding ExV8-TH THs were sown in a climate-controlled greenhouse in 10 cm × 10 cm pots, with nine pots of each genotype. Each DH line was always paired adjacently to its respective TH. Initially, three seeds per pot were sown in soil, and each pot was thinned to one seedling per pot at 7 days after sowing (das). An alpha-lattice design with two randomised replicates was applied, in each case using 20 blocks comprising 13 pairs of accessions that were each sown and harvested on the same day. The parental lines ‘Express 617’ and ‘V8’ were included as controls. At 14 das, the time point when the epicotyls generally started to appear, hypocotyl length was measured on five plants per genotype without removing the seedlings. The shoot fresh weight of the nine remaining plants of each genotype was measured at 28 das by weighing directly after harvesting, and dry weight was measured after oven-drying. Before drying, leaf area at 28 das was measured on two plants per genotype. The leaves were scanned using a Hewlett Packard Scanjet 5400c flatbed scanner and the images were processed using ImageJ 1.37v (http://rsb.info.nih.gob/ij/) to calculate the leaf area. Specific leaf weight was calculated as the proportion of leaf area versus leaf dry weight.
In Göttingen, a similar greenhouse trial with four replications was performed with the 250 ExR53-DH lines and their corresponding ExR53-TH THs, using ‘Express 617’, ‘R53’, the ExR53-F1 and the hybrid variety ‘Elektra’ as controls. In this experiment, the shoot fresh weight at 29 das was measured for estimation of QTL for seedling biomass and biomass heterosis. The different genotypes were sown in square plots consisting of nine pots (7 cm × 7 cm). The multi-array plots were organised on 12 tables. To adjust for light and temperature differences in the greenhouse, a 26 × 10 alpha lattice design (Patterson and Williams 1976) was applied. Each of the checks, ‘Express 617’ and ‘R53’ for the DH lines and the ExR53-F1 and ‘Elektra’ for the THs were replicated five times within the lattice. The DH lines and their corresponding hybrids followed the same alpha design but were grown on different tables in order to exclude competition between them. A single row of pots, sown with ‘Express 617’ or ‘Elektra’, was placed around the borders of each table with DH-lines or hybrids, respectively, to avoid border effects. Two seeds were sown in each pot, and at the first leaf stage one of the plants was removed to minimise competition for the remaining plant. The harvest was done in 1 day for all genotypes of a replication.
Field trials
The ExV8-DH/TH and ExR53-DH/TH populations were evaluated together in field trials at four locations in Germany in the growing seasons 2005–2006 (harvest in July 2006) and 2006–2007 (harvest in July 2007). The locations used were Rauischholzhausen and Grund-Schwalheim in Middle Hesse, along with Reinshof and Einbeck in Lower Saxony. At each location, the ExV8 and ExR53 materials were grown in adjacent trials. The trials were performed using an alpha-lattice design with 26 blocks of 10 plots each. Harvested plot areas varied from 10 to 13.5 m2 at the different locations. Randomisation was conducted using ALPHA+© ver. 2.4 (1998) from CSIRO/Biomathematics and Statistics Scotland. Each block was divided into two halves that were sown parallel to each other. One half of each block contained ten plots of DH lines, whereas the opposite half of the block contained the respective plots of hybrids derived from the same ten DH lines. No replications were possible except for the standards, which were included in five replicates. As a consequence, locations became replicates in the analysis and genotype–location interaction effects could not be estimated.
Plant height at the end of flowering and thousand seed mass (TSM) were measured in both sets of DH and TH populations, while plot yields were converted to yield potential in dt per ha with correction for a seed water content of 9%. Seeds per silique and siliques per square decimetre were measured as additional yield parameters in the ExR53-DH/TH populations in the growing season 2005–2006, as described by Radoev et al. (2008).
Statistical analysis
PROC MIXED from SAS® 8.02 was employed to analyse the observation data and to obtain least square mean estimates of the trait values for each accession. Data from each population were pooled over repetitions in the respective greenhouse trials in Giessen and Göttingen, whereas the analysis of the field trials was conducted for each year individually as well as pooled over years for the respective populations. The model statement was arranged following Piepho et al. (2003) based on the alpha-lattice trial design. Accessions, locations (as replicates), years and accession-to-year interactions were treated as random effects. Variance components from PROC MIXED output were used to estimate the heritability (h 2) for each trait in each population.
Genetic mapping and QTL analysis
The genetic maps for the ExV8 and ExR53 DH populations, generated using AFLP (Vos et al. 1995) and simple sequence repeat (SSR) markers, have both been published previously (Basunanda et al. 2007 and Radoev et al. 2008, respectively). These publications both used the N01-N19 chromosome nomenclature for B. napus (e.g. Parkin et al. 2005), whereby chromosomes N01-N10 correspond to the A-genome chromosomes A01-A10 derived from B. rapa, and N11-N19 to the C-genome chromosomes C01-C09 from B. oleracea, according to the nomenclature suggested by the Multinational Brassica Genome Project (see www.brassica.info). Since the amphidiploid B. napus genome has extensive duplications, only markers whose segregation did not show excessive distortion were included in the maps. Due to the large number of randomly distributed AFLP and SSR markers, and because unpublished maps of both populations with considerably higher numbers of markers had a similar overall map length, the maps were expected to provide a high coverage of the genome.
To assist in the alignment of the two maps, where possible we preferentially used SSR markers that were polymorphic in both populations. QTL analysis was conducted using the freely available software QTLNetwork 2.0 (Yang et al. 2007), with QTL detection probability threshold levels set to the equivalent of 0.005. For all traits, QTL were analysed using the per se data from the two DH populations, the backcross data from the two TH populations and the mid-parent heterosis (MPH) data calculated from trait values of each TH, its corresponding DH line parent and ‘Express 617’. Since the genetic maps were obtained from the respective DH populations, the QTL analysis of all three sets of data needed to be analysed using the _ Population DH option. As a result, the genetic architecture of each dataset needs to be taken into account when interpreting the genetic effects measured by the software in each case. The data from the DH lines give a direct estimate of the additive genetic effects at each QTL. On the other hand, the estimated additive effects of QTL from the TH (backcross) dataset only account for half of the difference between additive and dominant genetic effects, whereas the MPH dataset estimates half (negative) of the total dominance effects of each QTL. For details on the statistical methods used for the QTL mapping, and on the calculation and interpretation of the genetic effects with the given population structures, see Radoev et al. (2008).
Epistatic interactions among loci in all datasets from ExV8 and ExR53 were also estimated using QTLNetwork 2.0. In the case of epistasis, the estimated effect in the DH population is the additive × additive genetic interaction. The effects calculated in the other two data sets represent complex mixtures of all possible epistatic interactions: additive × additive (aa), additive × dominance (ad) and dominance × dominance (dd) interactions. If two loci A and B are considered, then the genetic effect in the testcross population represents aa AB + dd AB − ad AB − ad BA , whereas the effects estimated with MPH data are dd AB − aa AB − ad AB − ad BA .
Results
Analysis of genetic variance
Significant differences were found among accessions in each dataset for all traits. Variance components for the respective DH and TH populations, along with the estimated heritability estimates for all traits measured in multiple environments, are given in Table 1. Moderate-to-high heritability was observed in both DH populations for plant height, yield and TSM and seed yield, and for silique traits in ExR53-DH. In contrast, the seedling biomass traits showed relatively low heritabilities. As expected, the genotypic variance and heritabilities for almost all traits were lower in the two TH populations than in the corresponding DH populations, due to the common genetic background contributed to each backcrossed TH by the recurrent parent ‘MSL-Express’.
Average MPH levels for each trait in the two populations of THs are shown in Table 2 along with levels of MPH in the respective F1 from the two crosses. Significant MPH was observed for biomass traits even during early seedling development, with hypocotyl length, leaf weight, leaf area and shoot weight also showing best-parent heterosis (BPH) in ExV8 and shoot fresh weight also showing BPH in ExR53. The F1 offspring from both ExV8 and ExR53 exhibited significant MPH for seed yield, with the ExV8-F1 also exhibiting positive BPH in 2007. The ExR53-F1 showed BPH for yield in 2006 and plant height in 2007. Both the ExV8-TH and ExR53-TH populations also showed mean MPH for seed yield, whereas MPH for TSM was either non-significant or negative for the F1 offspring and over the population means. As described previously by Radoev et al. (2008), mean MPH and BPH were also measured for seeds per silique traits in the ExR53-TH population. Mean population yields were lower in the ExR53 materials than in the corresponding ExV8-DH and TH populations, presumably due to the unadapted nature of ‘R53’.
Detection of main effect QTL
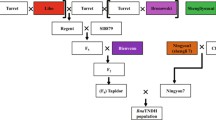
The comparative positions of all significant main-effect QTL detected in the DH, TH and MPH datasets from the ExV8 and ExR53 populations are given in Fig. 1. Full details of QTL peak positions, flanking markers, intervals, effects and the explained phenotypic variance of the complete set of QTL are provided in Supplementary Table 1. Noticeable clustering of QTL for different seedling and yield traits from the DH, TH and MPH datasets can be observed in both the ExV8 and ExR53 genetic maps, and the positions of many of these hotspots correspond between the two maps. Seedling biomass traits had less detected QTL than traits observed in the field trials, reflecting the relatively weak intergenotypic variation detected in the earlier stage of development. The phenotypic variances explained by QTL calculated from the DH populations tended to be higher than those from the MPH datasets, suggesting stronger influences of additive-acting than dominance-acting loci. QTL detected in the TH populations generally showed a higher contributed variance than those detected in the DH and MPH datasets. This was expected since the estimates of variance obtained from the TH (backcross) data contain both additive and dominance components.
Comparative map positions of main effect QTL for seedling and seed yield-related traits in DH and backcrossed TH populations derived from the crosses ‘Express 617’ × ‘V8’ (ExV8) and ‘Express 617’ × ‘R53’ (ExR53) along with MPH data from ExV8 and ExR53, respectively. QTL for silique traits were calculated previously by Radoev et al. (2008). To facilitate comparisons with previous publications the chromosome nomenclature follows the N01-N19 nomenclature for B. napus, with the A and C genome nomenclature suggested by the Multinational Brassica Genome Project (see www.brassica.info) given in brackets. White (DH data), grey (TH data) and black (MPH data) bars show the 1-logarithm of odds (LOD) intervals for each QTL. The prefix to each QTL name indicates the dataset from which the QTL was calculated, whereas the traits are abbreviated as follows: y yield, TSM thousand seed mass, ph plant height at flowering, slpa siliques per unit area, spsl seeds per silique, 06 2006 field trials, 07 2007 field trials, lfw leaf fresh weight, sfw shoot fresh weight, hcl hypocotyl length, la leaf area
Clusters of heterosis-relevant QTL
Clusters of QTL at corresponding positions in both ‘Express 617’ × ‘V8’ and ‘Express 617’ × ‘R53’ were observed on N01/N11, N03/N13, N05, N08 (presumably LG10 in ‘Express 617’ × ‘R53’ map), N07/N16 and N09/N19 and N10, whereby the pairs represent chromosomes with extensive homoeology (Udall et al. 2006).
Of particular interest were a number of chromosome regions that showed colocalisation between the ExV8 and ExR53 maps of heterosis-related QTL for both seedling and yield traits. For example, a QTL for heterosis for plant height at flowering on chromosome N11 in ExV8 co-localised with QTL for fresh leaf weight in the ExV8-DH and ExV8-TH populations, respectively, while the corresponding chromosome region also harboured a QTL for heterosis in seeds per silique in ExR53 along with QTL influencing siliques per unit area, seeds per silique, TSM and fresh shoot weight in the ExR53-DH population, and also QTL for fresh shoot weight in the ExR53-TH population. Moreover, this QTL may be homoeologous to a region of chromosome N01 containing QTL for plant height in both the ExV8-DH and ExR53-DH populations. A similar hotspot of putative homoeologous, heterosis-related QTL for different traits was observed on chromosomes N03 and N13. In this case, QTL for yield heterosis in both ExV8 (N03 and N13) and ExR53 (N13) clustered at corresponding positions with QTL for heterosis in TSM (ExR53-MPH), for TSM in the ExR53-TH population, for siliques per unit area in the ExR53-DH population and for yield and fresh shoot weight, respectively, in the ExR53-DH population. A QTL for heterosis in plant height at flowering on chromosome N05 in ExV8 overlapped with QTL for seedling leaf area and also for seed yield in the ExV8-TH population, along with QTL for seed yield, seedling hypocotyl height and specific leaf weight in the ExV8-DH population. Two SSR markers within this QTL showed that the corresponding region also influences siliques per unit area and TSM in the ExR53-DH population and siliques per unit area in the ExR53-TH population. A yield heterosis QTL on chromosome N12 in ExR53 co-localised with QTL for yield, TSM and siliques per unit area in the ExR53-DH population, and a common SSR marker showed the homoeology of this QTL cluster to a QTL for plant height in the ExV8-TH population. A further interesting QTL hotspot was seen on N19, where putatively corresponding QTL for TSM heterosis in both ExV8 and ExR53 overlapped with QTL for heterosis in siliques per unit area that also influenced the same trait in the ExR53-DH population.
Chromosomes N07 and N16 are known in B. napus to contain homoeologous stretches. On N07, we observed overlapping QTL for heterosis in TSM in ExV8, and also for TSM in the ExV8-DH, ExR53-DH, ExV8-TH and ExR53-TH populations. An SSR marker within this QTL in ExV8 was also localised in QTL for seed yield in the ExR53-DH population and for siliques per unit area in the ExR53-TH population. Chromosome N16 also harboured a QTL for heterosis in TSM in ExV8 that could potentially be homoeologous to a corresponding QTL on N07. This region also contained further overlapping QTL for seed yield heterosis in ExV8, and for TSM and plant height in both the ExV8-DH and ExV8-TH populations, whereas the corresponding position also exhibited a QTL for TSM in the ExR53-DH population. Putatively corresponding QTL for heterosis in TSM were also obtained on N19 in both ExV8 and ExR53, and in ExR53 this QTL overlapped with a QTL for heterosis in siliques per unit area, and also influenced the same trait in the DH dataset. This region of N19 could potentially be homoeologous to the heterosis-related QTL cluster on N09, although no homoeologous markers were identified to substantiate this hypothesis.
Altogether, these examples provide substantial indications for clustering of QTL involved in numerous different traits in different developmental stages (seedling development, onset of flowering and seed-set, respectively) and in different environments (independent greenhouse experiments, multi-location field experiments over two different years). On chromosomes N01, N03, N05, N07, N11, N12, N13 and N16, the correspondence of the QTL cluster between the ExV8 and ExR53 genetic maps was corroborated by common SSR markers in the QTL regions, whereas in the case of N01 and N11 the corroboration was possible over homoeologous chromosomes in the two genetic maps.
QTL effects and digenic epistatic interactions
As found previously by Radoev et al. (2008) for the first year of field trials in ExR53, all levels of dominance from partial dominance through to overdominance were found to play a role in the expression of heterosis (data not shown). Complete dominance or overdominance was observed for seed yield, whereas most other traits showed mainly partial dominance. A summary of all first-order epistatic interactions calculated by the QTLNetwork software is given in Supplementary Table 2. The number of interactions detected by QTLNetwork was somewhat higher than the number of epistatic interactions detected by Radoev et al. (2008) using the QTL Mapper software (Wang et al. 1999) and the 2006 field data from ExR53. However, the general trend of epistasis detection shown by both software packages was the same. For both ExV8 and ExR53, a large number of digenic interactions were detected, confirming the important role of epistasis in the expression of complex trait including heterosis. No Type I epistasis (interactions between two main-effect QTL) was detected, with the vast majority (97%) of digenic interactions being Type III epistasis (interactions between two loci with no main-effect QTL). Only 3% of the interactions were Type II epistasis (interactions between a main-effect QTL and a locus with no main-effect QTL). We confirm the finding of Radoev et al. (2008) that epistasis together with all levels of dominance from partial to overdominance is responsible for the expression of heterosis in oilseed rape.
Discussion
Heterosis and the related phenomenon of inbreeding depression are essential components of fitness that are quantitatively influenced by numerous gene loci and in particular by the environment (Mather and Jinks 1982). Due to the complex quantitative nature of heterosis, its analysis in both model and crop plants has generally been performed using QTL analysis methods, particularly for the model species rice, the classical hybrid crop maize and more recently for the model crucifer Arabidopsis. A major obstacle to the performance of such studies in an annual field crop species like winter oilseed rape is the difficulty in generating and testing large plant populations with an allelic structure that is suitable for the analysis of additive, dominance and epistatic gene action. This study generated large corresponding DH and backcrossed TH populations, specifically for the purpose of analysing heterosis for seedling biomass and yield traits. In this respect, the data presented represent a significant advance in heterosis research in winter oilseed rape. Furthermore, the use of common genetic markers allowed in many cases the alignment of QTL between different maps, which adds considerable value in comparison to QTL data from a single biparental cross.
A key underlying outcome of many previous QTL studies investigating the expression of heterosis was the importance of epistatic interactions. Although epistasis among main-effect QTL for heterosis is often not detectable (e.g. Xiao et al. 1995; Mihaljevic et al. 2005) or at least very low (Radoev et al. 2008), digenic epistatic interactions that do not necessarily involve main-effect loci have been found to play a key role in the expression of heterosis in rice (Yu et al. 1997; Li et al. 2001; Luo et al. 2001; Li et al. 2008), maize (Reif et al. 2009), Arabidopsis (Kusterer et al. 2007; Melchinger et al. 2007) and also oilseed rape (Radoev et al. 2008). In Arabidopsis, additive × additive epistatic interactions with the genetic background were found by Melchinger et al. (2007) to be particularly influential on biomass heterosis.
In addition to confirming the key role of epistatic interactions in determining heterosis in B. napus, we identified numerous genome regions with prominent clusters of heterosis-related QTL that appeared to coincide in two different genetic backgrounds. In many cases the QTL alignment could be confirmed by common SSR markers. In other cases, this might be presumed, because the fact that the two maps share a common mapping parent increases the probability that putatively corresponding QTL hotspots on common chromosomes in the two maps indeed represent the same genetic loci. This is particularly the case when QTL for the same or related traits appear at similar positions.
Because key yield components like silique traits and seed weight have a close interaction with each other and with seed yield, and due to the contribution of seed storage reserves to germination and seedling development there is also an undeniable interaction of seed weight with seedling development. Therefore, pleiotropy among QTL for these traits is not altogether unexpected. On the other hand, in many cases QTL appeared to co-localise for traits that at first sight do not appear to be developmentally related, for example seedling biomass traits, plant height at flowering and seed yield. Interestingly, correlation analysis of MPH data from ExV8 revealed weak but significant correlations between heterosis for shoot weight and heterosis for seed yield, and hybrids with the lowest and highest levels of heterosis for shoot weight also showed significantly lower and higher heterosis for seed yield, respectively (data not shown). In plants, the ovary and consequently the silique are phylogenetically related to leaves; hence, it is possible that regulatory factors involved in heterosis for leaf development in seedlings may also contribute to heterosis during ovary and silique development.
The coincidence of QTL between ExV8 and ExR53 is not altogether unexpected, given that these two populations share the common parent ‘Express 617’. On the other hand, the QTL clusters observed in this study also show striking parallels with the positions of QTL clusters for various yield-related traits that were reported in previous studies, in completely unrelated populations of THs. With the exception only of chromosome N08, all of the B. napus chromosomes where we observed heterosis-related QTL clusters were also found by Quijada et al. (2006) and Udall et al. (2006) to carry QTL clusters influencing yield-related traits in different DH and/or testcross populations. Chen et al. (2007) also identified yield-related QTL clusters on the same chromosomes using pairs of intercrossed homozygous lines from a DH mapping population. Although difficult to prove in the absence of a true consensus map or completed genome sequence for B. napus, the agreement between these results from very different studies and materials suggests the presence of key loci involved in expression of heterosis and yield. Some of these loci also appear to be involved in heterosis for seedling biomass traits. A feasible explanation is that these heterosis-related QTL clusters might represent key regulatory loci that infer a significant epistatic influence on gene expression throughout the plant life cycle. Interacting gene expression networks and their underlying transcription factors are thought to play an important role in heterosis (e.g. Meyer et al. 2007), so that the presence of common regulatory QTL is not altogether unexpected. On the other hand, the prevalence of Type III epistasis suggests complex higher-order interactions among genes that alone do not contribute significantly to main-effect QTL. The co-localisation of per se QTL for various yield-related traits with heterosis-related QTL indicates that regulatory loci might also be contributing to fixed heterosis in the highly duplicated B. napus genome. Such hotspots might harbour genes involved in transcriptional or post-transcriptional regulation of heterosis (including fixed heterosis) for different traits throughout the plant life cycle, including a significant overall influence on heterosis for seed yield.
QTL hotspots for heterosis in biomass traits and metabolites, respectively, were reported recently in Arabidopsis by Meyer et al. (2009) and Lisec et al. (2009). Significant clusters of heterotic metabolite QTL were observed by Lisec et al. (2009) at the bottom of Arabidopsis chromosome 1, the bottom of chromosome 3 and the top of chromosome 4. The latter two regions coincided with significant clusters of per se and heterotic biomass QTL in the study of Meyer et al. (2009), whereas the cluster on chromosome 4 also contained multiple per se metabolite QTL (Lisec et al. 2008) and was also involved in digenic epistatic interactions (Meyer et al. 2009). Together these results suggest the presence of at least one key regulatory factor that influences heterosis in multiple pathways contributing to metabolite biosynthesis and biomass accumulation. This is consistent with our findings, and after identification of the responsible gene or genes in Arabidopsis their duplicated orthologs in B. napus will be key candidates to elucidate the control of heterosis in the polyploid crop species.
On the other hand, the investigation of trans-acting regulatory loci corresponding to these hotspots by expression QTL analysis (eQTL; Darvasi 2003; Gibson and Weir 2005) or gene network co-expression studies (Zhang and Horvath 2005; Langfelder and Horvath 2007; Zhu et al. 2007) represents a promising parallel approach towards identification or confirmation of potential candidate genes. Since we could demonstrate that heterosis is manifested at a very early stage in B. napus seedlings, and that in some cases the same regulatory loci may be involved in the expression of heterosis for seedling and yield traits, this opens the possibility to use seedlings as a promising controlled system for eQTL analysis of yield-relevant gene expression in winter oilseed rape. We are currently performing eQTL analysis in seedlings from the ExV8-TH population, using next-generation sequencing-based digital gene expression profiling.
References
Basunanda P, Spiller TH, Hasan M, Gehringer A, Schondelmaier J, Lühs W, Friedt W, Snowdon RJ (2007) Marker-assisted increase of genetic diversity in a double-low seed quality winter oilseed rape genetic background. Plant Breed 126:581–587
Chen W, Zhang Y, Liu XP, Chen BY, Tu JX, Fu TD (2007) Detection of QTL for six yield-related traits in oilseed rape (Brassica napus) using DH and immortalized F2 populations. Theor Appl Genet 115:849–858
Darvasi A (2003) Genomics—gene expression meets genetics. Nature 422(6929):269–270
Gibson G, Weir B (2005) The quantitative genetics of transcription. Trends Genet 21:616–623
Kusterer B, Piepho HP, Utz HF, Schön CC, Muminovic J, Meyer RC, Altmann T, Melchinger AE (2007) Heterosis for biomass-related traits in Arabidopsis investigated by quantitative trait loci analysis of the triple testcross design with recombinant inbred lines. Genetics 177:1839–1850
Langfelder P, Horvath S (2007) Eigengene networks for studying the relationships between co-expression modules. BMC Syst Biol 1:54
Li ZK, Luo LJ, Mei HW, Wang DL, Shu QY, Tabien R, Zhong DB, Ying CS, Stansel JW, Khush GS, Paterson AH (2001) Overdominant epistatic loci are the primary genetic basis of inbreeding depression and heterosis in rice I. Biomass and grain yield. Genetics 158:1737–1753
Li L, Lu K, Chen Z, Mu T, Hu Z, Li X (2008) Dominance, overdominance and epistasis condition the heterosis in two heterotic rice hybrids. Genetics 180:1725–1742
Lisec J, Meyer RC, Steinfath M, Redestig H, Becher M, Witucka-Wall H, Fiehn O, Törjék O, Selbig J, Altmann T, Willmitzer L (2008) Identification of metabolic and biomass QTL in Arabidopsis thaliana in a parallel analysis of RIL and IL populations. Plant J 53:960–972
Lisec J, Steinfath M, Meyer RC, Selbig J, Melchinger AE, Willmitzer L, Altmann T (2009) Identification of heterotic metabolite QTL in Arabidopsis thaliana RIL and IL populations. Plant J. doi:10.1111/j.1365-313X.2009.03910.x
Luo LJ, Li Z-K, Mei HW, Shu QY, Tabiel R, Zhong DB, Ying CS, Stansel JW, Khush GS, Paterson AH (2001) Overdominant epistatic loci are the primary genetic basis of inbreeding depression and heterosis in rice II. Grain yield components. Genetics 158:1755–1771
Mather K, Jinks JL (1982) Biometrical genetics, 3rd edn. Chapman and Hall, London, 396 pp
Melchinger AE, Piepho H-P, Utz HF, Muminovi J, Wegenast T, Törjék O, Altmann T, Kusterer B (2007) Genetic basis of heterosis for growth-related traits in Arabidopsis investigated by testcross progenies of near-isogenic lines reveals a significant role of epistasis. Genetics 177:1827–1837
Meyer RC, Torjek O, Becher M, Altmann T (2004) Heterosis of biomass production in Arabidopsis. Establishment during early development. Plant Physiol 134:1813–1823
Meyer S, Pospisil H, Scholten S (2007) Heterosis associated gene expression in maize embryos 6 days after fertilization exhibits additive, dominant and overdominant pattern. Plant Mol Biol 63:381–392
Meyer RC, Kusterer B, Lisec J, Steinfath M, Becher M, Scharr H, Melchinger AE, Selbig J, Schurr U, Willmitzer L, Altmann T (2009) QTL analysis of early stage heterosis for biomass in Arabidopsis. Theor Appl Genet. doi:10.1007/s00122-009-1074-6
Mihaljevic R, Utz HF, Melchinger AE (2005) No evidence for epistasis in hybrid and per se performance of elite European flint maize inbreds from generation means and QTL analyses. Crop Sci 45:2605–2613
Parkin IA, Gulden SM, Sharpe AG, Lukens L, Trick M, Osborn TC, Lydiate DJ (2005) Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics 171:765–781
Patterson HD, Williams ER (1976) A new class of resolvable incomplete block designs. Biometrika 63:83–92
Piepho HP, Büchse A, Emrich K (2003) A hitchhiker’s guide to mixed models for randomized experiments. J Agron Crop Sci 189:310–322
Quijada PA, Udall JA, Lambert B, Osborn TC (2006) Quantitative trait analysis of seed yield and other complex traits in hybrid spring rapeseed (Brassica napus L.): 1. Identification of genomic regions from winter germplasm. Theor Appl Genet 113:549–561
Radoev M, Becker HC, Ecke W (2008) Genetic analysis of heterosis for yield and yield components in rapeseed (Brassica napus L.) by quantitative trait locus mapping. Genetics 179:1547–1558
Reif JC, Kusterer B, Piepho H-P, Meyer RC, Altmann T, Schon CC, Melchinger AE (2009) Unraveling epistasis with triple testcross progenies of near-isogenic lines. Genetics 181:247–257
Udall JA, Quijada PA, Lambert B, Osborn TC (2006) Quantitative trait analysis of seed yield and other complex traits in hybrid spring rapeseed (Brassica napus L.): 2. Identification of alleles from unadapted germplasm. Theor Appl Genet 113:597–609
Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Wang DL, Zhu J, Li ZKL, Paterson AH (1999) Mapping QTL with epistatic effects and QTL x environment interactions by mixed linear model approaches. Theor Appl Genet 99:1255–1264
Xiao J, Li J, Yuan L, Tanksley SD (1995) Dominance is the major genetic basis of heterosis in rice as revealed by QTL analysis using molecular markers. Genetics 140:745–754
Yang J, Zhu J, Williams RW (2007) Mapping the genetic architecture of complex traits in experimental populations. Bioinformatics 23:1527–1536
Yu SB, Li JX, Xu CG, Tan YF, Gao YJ, Li XH, Zhang Q, Maroof MAS (1997) Importance of epistasis as the genetic basis of heterosis in an elite rice hybrid. Proc Natl Acad Sci USA 94:9226–9231
Zhang B, Horvath S (2005) A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol 4: No. 1, Article 17
Zhu J, Wiener MC, Zhang C, Fridman A, Minch E et al (2007) Increasing the power to detect causal associations by combining genotypic and expression data in segregating populations. PLoS Comput Biol 3:e69
Acknowledgments
This study was funded by the German Research Foundation (DFG) as part of the priority program SPP 1149 “Heterosis in Plants”. The authors thank KWS Saat AG and SW Seed GmbH for valuable assistance with the field evaluations, and Norddeutsche Pflanzenzucht H.-G. Lembke KG for providing the male-sterile tester. Mechthild Schwarte, Malte Luh, Liane Renno (University of Giessen), Angela Pretorius, Petra Freynhagen and Gerald Miotke (University of Göttingen) provided excellent technical assistance.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by C. Quiros.
Contribution to the special issue “Heterosis in Plants”.
Electronic supplementary material
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Basunanda, P., Radoev, M., Ecke, W. et al. Comparative mapping of quantitative trait loci involved in heterosis for seedling and yield traits in oilseed rape (Brassica napus L.). Theor Appl Genet 120, 271–281 (2010). https://doi.org/10.1007/s00122-009-1133-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-009-1133-z