Abstract
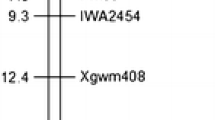
Powdery mildew, caused by Blumeria graminis f.sp. tritici, is one of the most important wheat diseases in many regions of the world. Triticum turgidum var. dicoccoides (2n = 4x = AABB), the progenitor of cultivated wheats, shows particular promises as a donor of useful genetic variation for several traits, including disease resistances. The wild emmer accession MG29896, resistant to powdery mildew, was backcrossed to the susceptible durum wheat cultivar Latino, and a set of backcross inbred lines (BC5F5) was produced. Genetic analysis of F3 populations from two resistant introgression lines (5BIL-29 × Latino and 5BIL-42 × Latino) indicated that the powdery mildew resistance is controlled by a single dominant gene. Molecular markers and the bulked segregant analysis were used to characterize and map the powdery mildew resistance. Five AFLP markers (XP43M32 (250), XP46M31 (410), XP41M37 (100), XP41M39 (250), XP39M32 (120)), three genomic SSR markers (Xcfd07, Xwmc75, Xgwm408) and one EST-derived SSR marker (BJ261635) were found to be linked to the resistance gene in 5BIL-29 and only the BJ261635 marker in 5BIL-42. By means of Chinese Spring nullisomic–tetrasomic, ditelosomic and deletion lines, the polymorphic markers and the resistance gene were assigned to chromosome bin 5BL6-0.29-0.76. These results indicated that the two lines had the same resistance gene and that the introgressed dicoccoides chromosome segment was longer (35.5 cM) in 5BIL-29 than that introgressed in 5BIL-42 (less than 1.5 cM). As no powdery mildew resistance gene has been reported on chromosome arm 5BL, the novel resistance gene derived from var. dicoccoides was designated Pm36. The 244 bp allele of BJ261635 in 5BIL-42 can be used for marker-assisted selection during the wheat resistance breeding process for facilitating gene pyramiding.



Similar content being viewed by others
References
Blanco A, Simeone R, Gadaleta A (2006) Detection of QTLs for grain protein content in durum wheat. Theor Appl Genet 112:1195–1204
Chantret N, Sourdille P, Roder M, Tavaud M, Bernard M, Doussinault G (2000) Location and mapping of the powdery mildew resistance gene MIRE and detection of a resistance QTL by bulked segregant analysis (BSA) with microsatellite in wheat. Theor Appl Genet 100:1217–1224
Devos KM, Miller T, Gale MD (1993) Comparative RFLP maps of the homoeologous group 2 chromosomes of wheat, rye and barley. Theor Appl Genet 85:784–792
Dilbirligi M, Erayman M, Sandhu D, Sidhu D, Gill KS (2004) Identification of wheat chromosomal regions containing expressed resistance genes. Genetics 166:461–481
Dinoor A, Eshed N, Ecker R, Gerechter-Amitai Z, Solel Z, Manisterski J, Anikster Y (1991) Fungal diseas of wild tetraploid wheat in a natural stand in northern Israel. Israel J Bot 40:481–495
Endo TR, Gill BS (1996) The deletion stocks of common wheat. J Hered 87:295–307
Eujayil I, Sorrels ME, Baum M, Volters P, Powell P (2002) Isolation of EST-derived microsatellite markers for genotyping the A and B genome of wheat. Theor Appl Genet 104:399–407
Faris JD, Haen KM, Gill BS (2000) Saturation mapping of a gene-rich recombination hot spot region in wheat. Genetics 154:823–835
Feldman M, Millet E (1993) Methodologies for identification, allocation and transfer of quantitative genes from wild emmer into cultivated wheat. In: Li ZS, Xin ZY (eds) Proceedings of the 8th international Wheat Genetics Symposium. China Agricultural Scientech Press, Beijing, pp 19–27
Feldman M, Liu B, Segal G, Abbo S, Levy AA, Vega JM (1997) Rapid elimination of low-copy DNA sequences in polyploid wheat: a possible mechanism for differentiation of homoeologous chromosomes. Genetics 147:1381–1387
Gill KS, Gill BS, Endo TR, Boyko EV (1996) Identification and high-density mapping of gene-rich regions in chromosome group 5 of wheat. Genetics 143:1001–1012
Gupta PK, Varshney RK, Sharma PC, Ramesh B (1999) Molecular markers and their applications in wheat breeding. Plant Breed 118:369–390
Gupta PK, Balyan HS, Edwards KJ, Isaac P, Korzun V, Röder M, Gautier MF, Joudrier P, Schlatter AR, Dubcovsky J, De la Pena RC, Khairallah M, Penner G, Hayden M.J, Sharp P, Keller B, Wang RCC, Hardouin JP, Jack P, Leroy P (2002) Genetic mapping of 66 new microsatellite (SSR) loci in bread wheat. Theor Appl Genet 105:413–422
Huang XQ, Röder MS (2004) Molecular mapping of powdery mildew resistance genes in wheat: a review. Euphytica 137:203–223
Joppa LR, Williams ND (1988) The Langdon durum disomic-substitutions and aneuploid analysis in tetraploid wheat. Genome 30:222–228
Keller M, Keller B, Schachermayr G, Winzeler M, Schid JE, Stamp P, Messmer MM (1999) Quantitative trait loci for resistance against powdery mildew in a segregating wheat × spelt population. Theor Appl Genet 98:903–912
Kosambi DD (1944) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Liu ZY, Sun QX, Ni ZF, Nevo E, Yang TM (2002) Molecular characterization of a novel powdery mildew resistance gene Pm30 in wheat originating from wild emmer. Euphytica 123:21–29
Lutz J, Hsam SLK, Limpert E, Zeller FJ (1995) Chromosomal location of powdery mildew resistance genes in Triticum aestivum L. (common wheat). 2. Genes Pm2 and Pm19 from Aegilops squarrosa L. Heredity 74:152–156
McIntosh RA, Baker EP (1970) Cytogenetic studies in wheat. IV. Chromosomal location and linkage studies involving the Pm2 locus for powdery mildew resistance. Euphytica 19:71–77
McIntosh RA, Yamazaky Y, Devos KM, Dubcovsky J, Rogers WJ, Appels R (2003) Catalogue of gene symbols for wheat. In: Pogna NE, Romanò M, Pogna EA, Galterio G (eds) Proceedings of 10th international wheat genetics symposium. Istituto Sperimentale Cerealicoltura, Rome
Michelmore RW, Paran I, Kesseli RV (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating population. Proc Natl Acad Sci USA 88:9828–9832
Mingeot D, Chantret N, Baret PV, Dekeyser A, Boukhatem N, Sourdille P, Doussinoult G, Jacquemin JM (2002) Mapping QTL involved in adult plant resistance to powdery mildew in the winter wheat line RE714 in two susceptible genetic background. Plant Breed 121:133–140
Miranda LM, Murphy JP, Leath S, Marshall DS (2006) Pm34: a new powdery mildew resistance gene transferred from Aegilops tauschii Coss. to common wheat (Triticum aestivum L.). Theor Appl Genet 113:1497–1504
Miranda LM, Murphy JP, Marshall DS, Cowger C, Leath S (2007) Chromosomal location of Pm35, a novel Aegilops tauschii derived powdery mildew resistance gene introgressed into common wheat (Triticum aestivum L.). Theor Appl Genet 114:1451–1456
Ozkan H, Levy AA, Feldman M (2001) Allopolyploidy-induced rapid genome evolution in the wheat (Aegilops–Triticum) group. Plant Cell 13:1735–1747
Reader M, Miller TE (1991) The introduction into bread wheat of major gene for resistance to powdery mildew from wild emmer wheat. Euphytica 53:57–60
Röder MS, Korzun V, Wendehake K, Plaschke J, Tixier MH, Leroy P, Ganal MW (1998) A microsatellite map of wheat. Genetics 149:2007–2023
Rong JK, Millet E, Manisterski J, Feldman M (2000) A new powdery mildew resistance gene: introgression from wild emmer into common wheat and RFLP-based mapping. Euphytica 115:121–126
Saari EE, Prescott JM (1975) A scale of appraising the foliar intensity of wheat diseases. Plant Dis Rep 59:377–380
Sears ER (1954) The aneuploids of common wheat. Agr Exp Stn Res Bull Univ Mo 572:1–58
Sears ER (1966) Nullisomic-tetrasomic combinations in hexaploid wheat. In: Riley R, Lewis KR (eds) Chromosome manipulations and plant genetics. Oliver and Boyd, Edinburgh, pp 20–45
Sears ER (1968) Relationship of chromosomes 2A, 2B and 2D with their rye homoeologoue. In: Finlay KW, Shepherd KW (eds) Proceedings of 3rd international wheat genetics symposium, Australian Academy Science, Canberra, pp 53–61
Sears ER, Sears LMS (1978) The telocentric chromosomes of common wheat. In: Ramanujam S (ed) Proceedings of 5th international wheat genetic symposium. Indian Society of Genetics and Plant Breeding, New Delhi, pp 389–407
Sharp PJ, Kreis M, Shewry PR, Gale MD (1988) Location of α-amylase sequences in wheat and its relatives. Theor Appl Genet 75:286–290
Somers DJ, Isaac P, Edwards K (2004) A high density microsatellite consensus map for bread wheat (Triticum aestivum L). Theor Appl Genet 109:1105–1114
Sorrels ME, La Rota M, Bermudes-Kandianis C, Greene RA et al (2003) Comparative DNA sequence analysis of wheat and rice genomes. Genome Res 13:1818–1827
Sourdille P, Cadalen T, Guyomarc’h H, Snape JW, Perretant MR, Charmet G, Boeuf C, Bernard S, Bernard M (2003) An update of the Courtot × Chinese Spring intervarietal molecular marker linkage map for the QTL detection of agronomic traits in wheat. Theor Appl Genet 106:530–538
Tanksley SD, Nelson JC (1996) Advanced backcross QTL analysis: a method for the simultaneous discovery and transfer of valuable QTLs from unadapted germplasm into elite breeding lines. Theor Appl Genet 92:191–203
Van Ooijen JW, Voorrips RE (2001) JoinMap v. 3, software in the calculation of genetic linkage maps. Plant Research International, Wageningen. http://www.Kyazma.nl/index,php/mc.JoinMap
Vos P, Hogers R, Bleeker M, Reijans M, van der Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 21:4407–4414
Wehrhahn C, Allard RW (1965) The detection and measurement of the effects of individual genes involved in the inheritance of a quantitative character in wheat. Genetics 51:09–119
Xie CJ, Sun QX, Ni ZF, Yang ZM, Nevo E, Fahima T (2003) Chromosomal location of a Triticum dicoccoides-derived powdery mildew resistance gene in common wheat by using microsatellite markers. Theor Appl Genet 106:341–345
Yahiaoui N, Srichumpa P, Dudler R, Keller B (2004) Genome analysis at different ploidy levels allows cloning of the powdery mildew resistance Pm3b from hexaploid wheat. Plant J 34:528–538
Zhu ZD, Zhou RH, Kong XY, Dong YC, Jia JZ (2005) Microsatellite markers linked to two genes conferring resistance to powdery mildew in common wheat introgressed from Triticum carthlicum accession PS5. Genome 48:585–590
Acknowledgments
This research was supported by a grant from the Ministero delle Politiche Agricole, Alimentari e Forestali, Italy, project “AMIFRUGAM”, and by the Ministero dell’Università e della Ricerca, Italy, project “AGROGEN”.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by I. Romagosa.
Rights and permissions
About this article
Cite this article
Blanco, A., Gadaleta, A., Cenci, A. et al. Molecular mapping of the novel powdery mildew resistance gene Pm36 introgressed from Triticum turgidum var. dicoccoides in durum wheat. Theor Appl Genet 117, 135–142 (2008). https://doi.org/10.1007/s00122-008-0760-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-008-0760-0