Abstract
Plant metabolism is perturbed by various abiotic stresses. As such the metabolic network of plants must be reconfigured under stress conditions in order to allow both the maintenance of metabolic homeostasis and the production of compounds that ameliorate the stress. The recent development and adoption of metabolomics and systems biology approaches enable us not only to gain a comprehensive overview, but also a detailed analysis of crucial components of the plant metabolic response to abiotic stresses. In this review we introduce the analytical methods used for plant metabolomics and describe their use in studies related to the metabolic response to water, temperature, light, nutrient limitation, ion and oxidative stresses. Both similarity and specificity of the metabolic responses against diverse abiotic stress are evaluated using data available in the literature. Classically discussed stress compounds such as proline, γ-amino butyrate and polyamines are reviewed, and the widespread importance of branched chain amino acid metabolism under stress condition is discussed. Finally, where possible, mechanistic insights into metabolic regulatory processes are discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
When plants face unfavourable growth conditions, abiotic stress retards plant growth and productivity. Under most abiotic stress conditions, plant metabolism is perturbed either because of inhibition of metabolic enzymes, shortage of substrate, excess demand for specific compounds or a combination of these factors and many other reasons. Therefore, the metabolic network must be reconfigured to maintain essential metabolism and to acclimate by adopting a new steady state in light of the prevailing stress conditions. This metabolic reprogramming is also necessary to meet the demand for anti-stress agents including compatible solutes, antioxidants and stress-responsive proteins. The accumulation of reactive oxygen species (ROS) is another problem causing oxidation and dysfunction of cellular components and in the worst case cell death. The optimisation of metabolic flux via the organellar electron transport chains is, moreover, crucial in order to dampen ROS production. Maintenance of the redox state in the cell is thus another important task to provide the reducing power required for ROS scavenging. Despite such important roles of metabolic regulation under stress conditions, our current understanding of this process is fragmented and far from complete.
Metabolomics is a powerful tool by which to gain a comprehensive perspective of how metabolic networks are regulated and has indeed been applied by many researches in recent years. It can, additionally, be used to elucidate the functions of genes as a tool in functional genomics and systems biology approaches. The term “metabolomics” is defined as comprehensive and quantitative analysis of all small molecules in a biological system [1]. The plant kingdom may contain between 200,000 and 1,000,000 metabolites, while for a single species the number may approach a few thousand (the estimate for Arabidopsis is ca. 5,000) [2–5]. Indeed the KNApSAck database (http://kanaya.naist.jp/KNApSAcK/) contains around 50,000 metabolite entries in plants described so far in the literature [6]. Due to the large variety of chemical structures and properties of small molecules, there is so far no single technique to identify and quantify all of them. Even the most comprehensive methods detect only between 1,000 and 2,000 molecular features [7, 8]. Several techniques including gas chromatography-mass spectrometry (GC-MS), liquid chromatography (LC)-MS, capillary electrophoresis (CE)-MS and nuclear magnetic resonance spectroscopy (NMR) are commonly used in plant metabolomics research. They are used sometimes in combination since they are largely complimentary with each independent method having preferential coverage of diverse types of metabolite. Here we briefly introduce the advantages and limitations of each method. Thereafter studies employing the metabolomic approach to dissect plant response to abiotic stresses will be discussed. Finally, using data already collected, we attempt to elucidate the general and stress-specific responses.
Techniques used for plant metabolomic research
Gas chromatography-mass spectrometry (GC-MS)
Gas chromatography-mass spectrometry is the most widely used technique for plant metabolomics research to date. Polar metabolites are derivatised to render them volatile and then separated by GC. Electron impact (EI) allows robust interfacing of GC with MS resulting in highly reproducible fragmentation patterns. For detection, time-of-flight (TOF)-MS has become the method of choice because of advantages including fast scan times, which give rise to either improved deconvolution or reduced run times for complex mixtures, and high mass accuracy. The crucial advantage of this technology is that it has long been used for metabolite profiling, and thus there are stable protocols for machine setup and maintenance, and chromatogram evaluation and interpretation [9–11]. The robustness of the protocol means that libraries of retention time and mass spectra data for standard compounds can be shared among laboratories [12]. There are several metabolite databases available including the NIST [13], FiehnLib [14] and Golm metabolic databases (GMD, [15]), which are useful tools for peak annotation. Additionally, the short running time and relatively low running cost are also strong advantages of GC-MS. However the use of GC-MS is limited for thermally stable volatile compounds, making the analysis of high molecular weight compounds (larger than 1 kDa) difficult. Due to these characteristics, GC-MS facilitates the identification and robust quantification of a few hundred metabolites in plant samples including sugars, sugar alcohols, amino acids, organic acids and polyamines, resulting in fairly comprehensive coverage of the central pathways of primary metabolism.
Liquid chromatography (LC)-MS
While GC has a limitation due to volatilisation of compounds, LC does not require prior sample treatment and separates the components in a liquid phase. The choice of columns, including reversed phase, ion exchange and hydrophobic interaction columns, provides the separation of metabolites based on different chemical properties. Therefore, LC has the potential to analyse a wide variety of metabolites in plants. The recent development of ultra-performance liquid chromatography (UPLC) makes the technique more powerful because of its higher resolution, sensitivity and throughput than conventional high-performance liquid chromatography (HPLC) [16]. Electrospray ionisation (ESI) is widely used for ionisation to connect LC and MS. Many types of MS including quadrupole (Q), TOF, qTOF, triple quadrupole (QqQ), ion trap (IT), linear trap quadrupole (LTQ)-Orbitrap and Fourier transform ion cyclotron resonance (FT-ICR)-MS are used depending on the sensitivity, mass-resolution and dynamic range required (see [17, 18] for the detail). The combination of these techniques allows us to identify and quantify a large variety of metabolites even if they have high molecular mass, great polarity and low thermostability. On the other hand the flexibility of the method also causes difficulty in establishing large mass spectral libraries for peak identification because of the instrument-type dependent retention time and mass spectra [19], and forces each research group to create its own “in-house” LC-MS reference library. That said, there are a number of websites that aid in mass-spectral analyses [20], and recent recommendations for metabolite reporting [7] should improve the transparency of the methods used by researchers. Furthermore, isotope labelling as a means of confirming the identity of peaks has recently been proposed and demonstrated to allow the identification of circa 1,000 metabolites using the FT-ICR-MS approach [8, 21]. To date, LC-MS is mainly used with a reverse phase column to analyse secondary metabolites because of its ability to separate compounds with similar structure and to detect a wide range of metabolites. However, it is worth noting that specialised protocols for determining phosphorylated intermediates, which are not readily detected by GC-MS, have also begun to be developed using this technology [22], as have methods for the comprehensive analysis of phytohormones [23].
Capillary electrophoresis (CE)-MS
Capillary electrophoresis separates polar and charged compounds on the basis of their charge-to-mass ratio. CE is able to separate a diverse range of chemical compounds and is a more powerful technique than LC with respect to separation efficiency [24, 25]. ESI is commonly used for ionisation as in LC-MS, with TOF-MS being the most commonly used detector in CE-MS-based metabolomics studies. This combination provides high mass accuracy and high resolution. The high scan speed of TOF-MS makes this instrument very suitable for full scan analyses in metabolomics. One of the unique properties of CE-MS is the small amount of sample required for analysis; only nanolitres of sample are introduced into the capillary. Together with high electric fields and short separation lengths, it can produce analysis within seconds. It also allows the metabolic analysis in volume-restricted samples. On the other hand, this leads to low concentration sensitivity requiring enrichment of metabolites within the samples [26]. Another drawback of CE is the poor migration time reproducibility and lack of reference libraries, which may only be partially overcome by the prediction of migration time [27]. Since CE and LC can both separate a large variety of metabolites via fundamentally different mechanisms, they are often used in combination to provide a wider coverage of metabolites [28–30]. That said, the use of CE-MS in plant studies remains relatively rare.
Nuclear magnetic resonance (NMR) spectroscopy
Nuclear magnetic resonance spectroscopy offers an entirely different analytical technique to that afforded by MS-based techniques being based on atomic interaction. In a strong magnetic field, atoms with non-zero magnetic moment including 1H, 13C, 14N, 15N and 31P absorb and re-emit electromagnetic radiation. The radiation is characterised by its frequency (chemical shift), intensity, fine structure and magnetic relaxation properties, all of which reflect the precise environment of the detected nucleus. Therefore, atoms in a molecule give a specific spectrum of radiation that can be used for identification and quantification of metabolites within a complex biological sample. The sensitivity of this method is much lower than that of MS-based techniques but the structural information content, reproducibility and quantitative aspects can be superior to them, and some journals require NMR spectra as the final proof of chemical structure [31, 32]. Furthermore the preparation of the sample is simple and even non-destructive measurement is possible. In vivo NMR can further generate kinetic measurements and examine metabolic responses on the same plant rather than on a set of similar plants [33]. The different subcellular pHs of the vacuole from the rest of the cell cause distinctive signals from an identical metabolite and thus allows quantification at the subcellular level [34, 35]. Thus analysing the metabolite composition of a tissue extract, determining the structure of a novel metabolite, demonstrating the existence of a particular metabolic pathway in vivo, isotope labelling experiment and localising the distribution of a metabolite in a tissue are all possible by NMR. For isotope labelling, NMR has the advantage of providing facile access to atomic level labelling, which is highly laborious in the case of MS methods yet can be essential in flux estimation [35]. However, the number of compounds that can be detected in a single analysis is limited to one to several dozen [36, 37]. These properties of NMR make it the ideal tool for broad-range profiling of abundant metabolites whilst studying changes in non-annotated profiles is highly useful for metabolite fingerprinting of extensive sample collections [38, 39].
Metabolomic studies of plant stress responses
Metabolomics is becoming increasingly common in plant physiology and biochemistry, and to date has been applied to a staggering number of conditions. Here we will attempt a synthesis of the most prominent studies dealing with plant stress; however, the reader is also referred to two previous reviews on this topic [40, 41]. In this section we independently review water stress, temperature stress, light stress, ionic stress, nutrient limitation and oxidative stress before discussing stress combinations. We describe the nature and symptoms of each stress, and then introduce several metabolomic studies with the main metabolic changes observed in each study and the conclusion drawn from the results. Following this survey we discuss commonalities and differences between the various stress responses.
Water stress
Water limitation is one of the major threats in crop production and this condition is projected to get considerably worse in coming decades [42]. For this reason considerable research effort has been expended to understand the response to this crucial and common stress. These studies have revealed an important role for metabolic regulation including regulation of photosynthesis and accumulation of osmolytes in the drought stress response [43, 44]. Urano et al. [28] reported metabolomic changes in Arabidopsis leaves under drought condition. The accumulation of many metabolites was observed, including amino acids such as proline, raffinose family oligosaccharides, γ-amino butyrate (GABA) and tricarboxylic acid (TCA) cycle metabolites, which are known to respond to drought stress in plants. The authors also investigated the nc3-2 mutant, which lacks the NCED3 gene involved in the dehydration-inducible biosynthesis of abscisic acid (ABA), in order to assess the effect of ABA in the metabolic response to drought stress. By combination with transcriptome analysis they clearly demonstrated that the ABA-dependent transcriptional regulation is responsible to the activation of metabolic pathways including branched chain amino acid, polyamine and proline biosynthesis, GABA shunt and saccharopin metabolism, but is not involved in the regulation of the raffinose biosynthetic pathway during dehydration.
In Arabidopsis research, drought tolerance is assessed predominantly under lethal conditions. However, in temperate climates, limited water availability rarely causes plant death but does restrict biomass and seed yield. Results of a recent elegant study experimentally demonstrated that the survival rate under lethal conditions does not predict superior growth performance and biomass yield gain under moderate drought [45], making this mild stress condition more important. Skirycz et al. [46] conducted metabolite profiling of Arabidopsis leaves that develop under mild osmotic stress. They revealed that the stress response measured in growing and mature leaves was largely distinct. Typical drought responses, namely accumulation of proline, erythritol and putrecine, were observed only in mature leaves, while many metabolites were decreased in expanding leaves, sharing the same tendency with transcriptional response. When we compared the data from the studies [28, 46], 24 metabolites were detected in both. The decrease of aspartate and increase of proline are the only two responses shared between mildly and severely desiccated leaves. Pronouncedly, amino acid metabolism responds in opposite ways; most amino acids were accumulated in severely desiccated leaves but decreased in mildly desiccated plants. These results highlight the variable response of plant metabolism in different developmental stages and degrees of desiccation. Metabolite profiling has additionally been carried out in crop species exposed to water stress conditions. Intriguingly, common changes in the levels of metabolites including branched chain amino acids were observed in wheat, barley and tomato [47–49].
Too much water, as occurs in situations such as flooding or water-logging of the rhizosphere, also causes problems because of the reduced oxygen availability (hypoxia/anoxia). Under anoxic conditions, ATP has to be produced by fermentation, resulting in cytosolic acidification and the accumulation of toxic products. van Dongen et al. [50] analysed metabolic responses in Arabidopsis roots under anoxic conditions. The accumulation of amino acids, alanine, proline and GABA, and the phosphoesters, glucose-6-phosphate and glycerol-3-phosphate, were observed as well as changes in the levels of minor sugars and various organic acids. When oxygen is decreased to 4 %, there is a general tendency for an increase in the levels of the intermediates both of sucrose degradation and the TCA cycle, and in the levels of most amino acids, whereas they are decreased when the oxygen further decreased to 1 %, indicating the inhibition and reactivation of metabolic activities. Together with the transcriptomic data showing a general downregulation of energy-consuming processes, the results demonstrated a large-scale reprogramming of metabolism under oxygen-limited conditions. Rocha et al. [51] examined the accumulation of alanine under anoxic conditions in Lotus japonicus, which is highly tolerant to water logging. In the roots of L. japonicus, succinate, alanine and the direct co-substrates for alanine synthesis, glutamate and GABA, were highly accumulated during water logging, whereas the majority of amino acids that are derived from TCA cycle intermediate decreased. The results are in agreement with the metabolic equilibriums that are expected to drive the metabolic flux from glycolysis, via alanine synthesis and oxoglutarate to succinate, which prevents the accumulation of pyruvate activating fermentation and leading to ATP production by succinyl-CoA ligase.
Temperature stress
Exposure to freezing environments leads to serious damage of the plant cell by ice formation and dysfunction of cellular membranes [52]. Many plant species increase freezing tolerance during exposure to non-freezing low temperature by a process known as “cold acclimation”. The molecular basis of this process has been extensively studied, and the contribution of particular metabolites including compatible solutes [53] and the transcriptional regulatory network has been elucidated [54, 55]. The first metabolomic studies of cold acclimation were performed by two groups in 2004. Cook et al. [56] compared metabolomic changes during cold acclimation in two ecotypes of Arabidopsis thaliana, Wassilewskija-2 (Ws-2) and Cape verde islands-1 (Cvi-1), which are relatively freezing tolerant and sensitive, respectively. The metabolome of Ws-2 plants was extensively altered in response to low temperature. Seventy-five percent of metabolites monitored were found to increase in cold-acclimated plants including metabolites known to increase in Arabidopsis plants upon exposure to low temperature, such as the amino acid proline and the sugars glucose, fructose, inositol, galactinol, raffinose and sucrose. They also found novel changes—namely the increase of trehalose, ascorbate, putrescine, citrulline and some TCA cycle intermediates. There was considerable overlap in the metabolite changes that occurred in the two ecotypes in response to low temperature; however, quantitative differences were evident. Kaplan et al. [57] conducted metabolome analysis of Arabidopsis over the time course following the shift to cold and heat conditions. Surprisingly the majority of heat shock responses were shared with cold shock including the increase of pool sizes of amino acids derived from pyruvate and oxaloacetate, polyamine precursors and compatible solutes. The results of this study were analysed together with following transcript profiling data by the same group [58], and revealed that the regulation of GABA shunt and proline accumulation under cold conditions are achieved by transcriptional and post-transcriptional manners, respectively. Gray and Heath [59] examined the effects of cold acclimation on the Arabidopsis metabolome using a non-targeted metabolic fingerprinting approach. It revealed a global reprogramming of metabolism as well as differential responses between the leaves that shifted to and those that developed in the cold. Hannah et al. [60] took advantage of the natural genetic variation of Arabidopsis to elucidate the function of metabolism in cold acclimation. Although there is no clear relationship between global metabolite changes and differences in acclimation capacity or differences between the accessions in acclimated freezing tolerance, the probable importance of central carbohydrate metabolism is indicated by the identification of glucose, fructose and sucrose among metabolites positively correlating to freezing tolerance. Espinoza et al. [61] analysed the effect of diurnal gene/metabolite regulation during cold acclimation by means of metabolomics and transcriptomics. Approximately 30 % of all analysed metabolites showed circadian oscillations in their pool size and low temperature affected the cyclic pattern of metabolite abundance. These results indicated that the interactions observed between circadian and cold regulation are likely highly relevant components of cold acclimation.
Metabolomics was also used to reveal the functions of specific genes in cold acclimation. In the study described above, Cook et al. [56] also investigated plants overexpressing CBF3, which is one of the C-repeat/dehydration responsive element-binding factor (CBF) transcriptional activators induced rapidly under low temperature conditions [62]. The metabolite profiles of non-acclimated CBF3 overexpressing lines were quite similar to those of the cold-acclimated Ws-2 ecotype, suggesting a prominent role for the CBF cold response pathway in configuring the low-temperature metabolome of Arabidopsis. Maruyama et al. [63] explored metabolic and transcript changes in Arabidopsis plants overexpressing CBF3/dehydration-responsive element binding protein (DREB)1A and another DREB protein DREB2A. They observed similar changes of metabolites in CBF3-overexpressing plants like Cook et al. [56] but DREB2A overexpression showed only a minor effect. The esk1 mutant is isolated as freezing tolerant without previous acclimation but the function of this gene had been unknown. Lugan et al. [64] tried to elucidate the basis of the freezing tolerance of esk1 by performing metabolomic analysis under various environmental conditions, namely cold, salinity and dehydration. Then the most specific metabolic responses to cold acclimation were not phenocopied by esk1 mutation. However, esk1 accumulated lower amount of Na+ in leaves than the wild type and its metabolic profile, and osmotic potential were only slightly impacted under dehydration stress. These results suggested that ESK1 could rather be involved in water homeostasis and as such highlighted the importance of cellular water status in stress tolerance.
Light stress
Light is a highly energetic substrate driving photosynthesis that can induce secondary destructive processes at the same time. Therefore, too high light irradiance represents an abiotic stress factor for plants. Wulff-Zottele et al. [65] conducted metabolite profiling of Arabidopsis leaves for 6 days after transition to high light. Generally, most of the metabolites of the glycolysis, TCA cycle and oxidative pentose phosphate pathway were altered in their content, indicating that plants exposed to high light undergo a metabolic shift and enhance the Calvin-Benson cycle to fix more carbon. In addition, elevation of glycine indicated the activation of photorespiratory pathways. Caldana et al. [66] investigated the early metabolic response against high light as a part of a more comprehensive study. The accumulation of the photorespiratory intermediates, glycine and glycolate, were observed in the early phase (5–60 min after transition). Interestingly the response during the mid phase (80–360 min) shares similar properties with low temperature treatment, which includes the accumulation of shikimate, phenylalanine and fructose, and the decrease of succinate; however, the physiological meaning of this overlap is currently unknown.
Not only the quantity but also the quality of light affects plant metabolism. In dense plant stands, such as crop fields or forests, individuals shade each other and create competition for light absorption [67]. Selective light absorption by the upper leaf layers leads to an enrichment of far-red wavelength [68], which induces excitation imbalances between photosystem II and I disturbing both the redox chemistry in the transport chain and its coordination with the Calvin-Benson cycle [69, 70]. For this reason, Bräutigam et al. [71] grew Arabidopsis plants under light, which preferentially excited either photosystem I (PSI light) or II (PSII light) and then transferred it to the other light condition to analyse how plants acclimate to the light quality shift. After long-term acclimation of 48 h, plants exhibited two distinct metabolic states. A PSI–II shift resulted in a decrease in primary products of photosynthesis, such as sugars, but an increase in important intermediates of subsequent metabolic pathways. By contrast, a PSII-I shift has no effect on the sugar pools but leads to general downregulation of many subsequent metabolites, including amino acids and organic acids. Each of the metabolites exhibited a different accumulation profile for establishing the final pool size, indicating high complexity by which the two metabolic states were achieved. Comprehensive analyses of these data alongside transcript profiles and other physiological data suggested that photosynthesis and metabolism were under the control of a binary combination of inputs from the thioredoxin and plastoquinone systems.
The dependency of plants upon sunlight also inevitably leads them into exposure to ultraviolet (UV) light, including in the wavelength range of 280–320 nm (UV-B). This wavelength potentially damages DNA, RNA and proteins, and additionally increases the production of free radicals [72, 73]. Kusano et al. [74] treated Arabidopsis plants with UV light and analysed the metabolic effect of UV light stress. Arabidopsis exhibits an apparent biphasic response to UV-B stress, characterised by major changes in the levels of primary metabolites, including ascorbate derivatives. By contrast, mid- to late-term responses were observed in the classically defined UV-B protectants, such as flavonoids and phenolics. The results suggested that in early stages of exposure to UV-B, the plant cell is ‘primed’ at the level of primary metabolism by a mechanism that involves reprogramming of the metabolism to efficiently divert carbon towards the aromatic amino acid precursors of the phenylpropanoid pathway. It also suggested the importance of ascorbate in the short-term response to UV-B. Further studies are, however, required to determine which of these metabolic changes are end responses to adapt to the enhanced exposure to UV-B and which are part of the perception-signalling relay, which alerts the plant cell that it needs to respond to the stress [75].
Ion stress
High levels of salinity in the soil hinder the growth and development of crops and cause serious problems for world food production [76]. High concentrations of NaCl may cause both hyperionic and hyperosmotic stress effects, which lead to a decline of turgor, disordered metabolism and the inhibition of uptake of essential ions, as well as other problems in plant cells [77, 78]. Gong et al. [79] conducted metabolite profiling of salt-treated Arabidopsis thaliana and its relative Thellungiella halophila (salt cress), which shows ‘extremophile’ characteristics manifested by extreme tolerance to a variety of abiotic stresses, among them low humidity, freezing and high salinity. Proline increased dramatically in both species as did inositols, hexoses and complex sugars. The concentrations of metabolites were often several-fold higher in Thellungiella and stress exacerbated the differences in some metabolites. Transcript analyses supported the metabolic results by suggesting that a Thellungiella is primed to anticipate such stresses. The difference in metabolites between Arabidopsis and Thellungiella under salt and osmotic stresses was more recently assessed for a broader range of metabolites [80]. Analysis of global physicochemical properties of metabolites revealed a shift from nonpolar to polar metabolites in both species but that this was much more pronounced in Thellungiella. Such a shift may contribute to keep the water potential during dehydration. Kim et al. [77] investigated the cellular level metabolic response using Arabidopsis T87 cultured cells. The results suggested that the methylation cycle for the supply of methyl groups, the phenylpropanoid pathway for lignin production and glycine betaine biosynthesis are synergetically induced as a short-term response against salt-stress treatment. The results also suggest the co-induction of glycolysis and sucrose metabolism as well as co-reduction of the methylation cycle as long-term responses to salt stress.
Due to the importance of salinity stress in agriculture, there are many metabolomic studies to assess the metabolic effect of salinity in a variety of crop and related plant species including tomato [40, 81], grapevine [82], poplar [83], sea lavender (Limonium latifolium, [84]) and rice [85]. Since these studies have been extensively reviewed in [40, 86], we focus here on three recent studies on legume species [87–89]. These recent studies took a functional genomic approach that integrated ionomic, transcriptomic and metabolomic analyses of the glycopyte model legume Lotus japonicus and other Lotus species subjected to long-term regimes of non-lethal levels of salinity. In Lotus japonicus the metabolic changes were characterised by a general increase in the steady-state levels of many amino acids, sugars and polyols, with a concurrent decrease in most organic acids [87]. The responses to salinity stress were compared between extremophile (L. creticus) and glycophytic (L. corniculatus and L. tenuis), but the metabolic responses were globally similar to each other [88]. These results suggest that, in contrast to Thellungiella, the metabolic pre-adaptation to salinity is not the major trait of L. creticus contributing to the extramophile phenotype. However, by comparing six species displaying different salt tolerances, they observed several genotype-specific features. One of them is the increase of asparagine levels in the more tolerant genotypes, suggesting that the roles of asparagine metabolism in supporting core nitrogen metabolism may play a role in tolerance [89].
Heavy metals such as cadmium (Cd), cesium (Cs), lead (Pb), zinc (Zn), nickel (Ni) and chromium (Cr) are major pollutants of the soil causing stress on plants. Even the essential nutrients including copper (Cu), iron (Fe) and manganese (Mn) can cause heavy metal stresses with inappropriate concentration. Generally heavy metals induce enzyme inhibition, cellular oxidation and metabolic perturbation, resulting in growth retardation and in extreme instances in plant death [90]. Jahangir et al. [91] analysed the effects of Cu, Fe and Mn on the metabolite levels of Brassica rapa, which is a known metal accumulator. Glucosinolates and hydroxycinnamic acids conjugated with malates as well as primary metabolites such as carbohydrates and amino acids were found to be the discriminating metabolites. Arabidopsis plants treated with Cd displayed increased levels of alanine, β-alanine, proline, serine, putrescine, sucrose and other metabolites with compatible solute-like properties, notably GABA, raffinose and trehalose [92]. This study also indicated that concentrations of antioxidants (α-tocopherol, campesterol, ß-sitosterol and isoflavone) also increased significantly. When taken together these data indicate an important role of antioxidant defences in the mechanisms of resistance to cadmium stress. Dubey et al. [93] conducted transcriptomic and metabolomic analysis of rice roots treated with Cr. Under these conditions proline accumulated to levels three-fold those of the control as did ornithine, which can be used in its synthesis. The content of several other metabolites including lactate, fructose, uracil and alanine increased following exposure to Cr stress; these were taken to suggest the modulation of the sucrose degradation pathway involving the three main fermentation pathways operating as a rescue mechanism when respiration is arrested. Further studies are however most likely warranted to gain a better understanding of the mechanisms underlying these changes.
Nutrient limitation
Nutrient starvation also dramatically affects plant growth and metabolism. Especially limitation of macronutrients, namely carbon (C), nitrogen (N), phosphorus (P) and sulphur (S), has direct effects on metabolism since most organic molecules comprise a combination of these elements. Changing environmental conditions continually alter the balance between C assimilation and utilisation. Even short periods of C starvation lead to an inhibition of growth, which is not immediately reversed when C becomes available again [94, 95]. Osuna et al. [96] investigated the metabolite profile of Arabidopsis seedlings in liquid culture under C starvation. In C-starved seedlings, as could be anticipated, carbohydrates, organic acids and other C-containing metabolites, including myo-inositol, raffinose, glycerate and fatty acids, decreased. Central amino acids (glutamine, glutamate, aspartate and alanine) and methionine, an S-containing amino acid, also decreased, indicating the inhibition of N and S assimilation, respectively. The increase of most other amino acids indicates that proteolysis has commenced. Most of these changes reverted rapidly after re-addition of sucrose into the media. Usadel et al. [97] took advantage of extended dark treatment to induce C starvation under more natural conditions in the Arabidopsis rosettes. Intriguingly, however, the changes in metabolite levels were mostly comparable to those observed in liquid culture seedlings [96]. The marked decrease of carbohydrates within the first 4 h of extended night indicates that the treatment induced C starvation very efficiently and that carbohydrates are starting to acutely limit metabolism. On the other hand, organic acids and other C-containing metabolites displayed a rather gradual decrease. The prolonged dark treatment induced severe C starvation and leaf senescence by the end of the experiment. The metabolite profile of Arabidopsis leaves subjected to prolonged darkness has been analysed in a series of studies to elucidate the metabolic bases of dark-induced senescence and the function of the mitochondrial alternative electron transport pathway during dark treatment [98–100]. Although a similar metabolic phenotype as the two studies described above [96, 97] was observed during the first few days of dark treatment, a subset of metabolites exhibits biphasic behaviour during prolonged exposure to darkness. This was particularly notable for some TCA cycle intermediates including fumarate, isocitrate, malate and succinate, which accumulated after 7 days of dark treatment despite decreasing during the first 3 days of treatment. Additionally accumulation of most amino acids including GABA became much more prominent. Metabolite profiles were also analysed in a range of mutants deficient in the genes involved in mitochondrial alternative electron transport mediated by the electron-transfer flavoprotein/electron-transfer flavoprotein:ubiquinone oxidoreductase (ETF/ETFQO) complex, namely ETFQO [98] and ETFβ [99] as well as enzymes involved in the provision of its substrates, namely IVDH, D2HGDH [101] and PSHX [100]. Although individual genotypes showed similar responses during the first 3 days of dark treatment, there are subtle differences in their metabolic complements at the end of the experiment, indicating an essential role of this alternative electron transport machinery during dark-induced starvation [99]. Further detailed analysis revealed that the ETF/ETFQO complex is involved in both the branched chain amino acids and the lysine catabolism pathways, and acts as an electron donor to the mitochondrial ubiquinol pool [100, 101]. These studies suggest that more integrative analysis of the role of all aspects of protein degradation and consequent remobilisation should be performed within the context of understanding metabolic responses to stress.
Nitrogen is required for the synthesis of nucleotides and amino acids, which are the building blocks of nucleic acids and proteins, and for the synthesis of phospholipids and many secondary metabolites that have diverse roles in signalling, structure and adaptation. The effect of N deficiency on the metabolite levels in tomato leaves were investigated by Urbanczyk-Wochniak and Fernie [102]. As would perhaps be expected, amino acid levels generally decreased under nitrogen deficiency. The level of 2-oxoglutarate, a key regulator of carbon and nitrogen interactions [103], decreased under N starvation as well as other TCA cycle intermediates including citrate, isocitrate, succinate, fumarate and malate. Tschoep et al. [104] analysed the effect of mild but sustained N limitation in Arabidopsis. Malate and fumarate levels were strongly decreased in low N conditions like in tomato leaves [102]. However, their rosette protein content was unaltered and total, and many individual amino acid levels increased compared with N-replete plants. The results revealed that Arabidopsis responds adaptively to low N condition. P is an essential component of intermediates in central and energy metabolism, signalling molecules and structural macromolecules like nucleic acids and phospholipids. Morcuende et al. [105] analysed the metabolite profile of Arabidopsis seedlings grown in liquid culture under P starvation. The levels of sugar phosphates were very low but metabolites further down in glycolysis, glycerate-3-phosphate, glycerate-2-phosphate and phosphoenolpyruvate, increased in P-deficient seedlings. Pi-deficient seedlings showed a marked accumulation of starch, sucrose and reducing sugars as well as a general increase of organic acids including citrate, fumarate, malate and oxoglutarate. The levels of most major amino acids did not alter or increased slightly, whereas those of several minor amino acids including the aromatic amino acids and histidine, arginine and threonine. Together with transcriptomic data, analysis of metabolites revealed that P deprivation leads to a shift towards the accumulation of carbohydrates, organic acids and amino acids. The effect of P starvation has also been studied on crop plants such as common bean and barley. Hernández et al. used metabolite profiling to assess the effect of P deficiency in the roots [106] and nodules [107] of the common bean. Most of the amino acids were increased in P-stressed roots. The accumulation of several sugars suggests that sugars may be partitioned preferentially to P-stressed roots to support the expression of P stress-induced genes. The reduced amounts of organic acids likely reflect exudation of these metabolites from the roots into the rhizosphere [106]. The metabolic response of P-starved nodules is in contrast to that of roots. Amino acids and other N-containing metabolites were decreased as well as sugars, while organic acids were accumulated in P-deficient nodules. Such a contrasting response may be due to the N deficiency in P-starved nodules in which the sole N supply from fixed N2 could be suppressed under environmental limitations such as P starvation [107]. Huang et al. [108] profiled metabolites from both shoots and roots of P-deficient barley. Severe P deficiency increased the levels of phosphorylated intermediates (glucose-6-P, fructose-6-P, inositol-1-P and glycerol-3-P) and organic acids (2-oxoglutarate, succinate, fumarate and malate). The results revealed that P-deficient plants modify carbohydrate metabolism initially to reduce P consumption and salvage P from small P-containing metabolites, which consequently reduce the levels of organic acid in the TCA cycle [108].
Sulphur is another macronutrient essential for the synthesis of the S-containing amino acids cysteine and methionine as well as a wide range of S-containing metabolites including glutathione. There are some metabolomic studies on the response to S starvation in Arabidopsis [109–112], and they are nicely summarised in Hoefgen et al. [113]. At the time course of S-stress response, two metabolic states can be distinguished. The short-term metabolic responses include the decrease of organic S-containing compounds on the S assimilation such as cysteine and glutathione, which leads to the accumulation of their precursor O-acetyl-serine (OAS) as well as serine, and to the subsequent re-channeling of the metabolic flow to glycine and tryptophan. Glucosinolate catabolism is activated to salvage S from it. As a long-term response the lipid contents and a S-containing molecule, S-adenosyl-methionine, decreased. Insufficient S supply leads to its disbalance with N and further to the alterations in C1 metabolism that link photorespiration, S assimilation and dumping of N [113]. Results of a very recent study on the Arabidopsis plants with modified OAS levels suggest the importance of this metabolite since OAS plays a signalling role for a specific part of the sulphate response as well as for the regulation of the transcript levels of a specific gene set irrespective of the sulphur status of the plants [114].
Potassium (K) is not a component of organic molecules but plays essential roles as a major cation in plants and as a cofactor of enzymes [115]. Armengaud et al. [116] used metabolite profiling to identify metabolic targets of K stress. Metabolite profiles of low-K Arabidopsis plants were characterised by a strong increase in the concentrations of soluble sugars (sucrose, fructose and glucose) and a slight net increase of total protein content and the overall amino acid level. Several basic or neutral amino acids accumulated during K deficiency, while acidic amino acids decreased. In addition a strong decrease of pyruvate and organic acids was recorded only in the roots but not in the shoots. They also measured enzyme activities and concluded that the primary effect of K deficiency induces an inhibition of glycolysis by the direct inhibition of enzymes [116].
Oxidative stress
Oxidative stress is a key underlying component of most abiotic stresses and a major limiting factor of plant growth in the field [117]. It occurs on the overproduction of reactive oxygen species (ROS) in plant cells when plant metabolism is perturbed by various stresses. This consequently leads to oxidative damages of cellular components such as DNA, proteins and lipids [118]. To cope with oxidative stress, the metabolic network of plant cells must be reconfigured either to bypass damaged enzymes or to support adaptive responses. In the study by Baxter et al. [119], heterotrophic Arabidopsis cells were treated with menadione, which enhances the ROS production via electron transport chains and changes in metabolite abundance, and 13C-labelling kinetics were quantified. The accumulation of sugar phosphates related to glycolysis and oxidative pentose phosphate pathways (OPPP) suggested the rerouting of glycolytic carbon flow into the OPPP possibly to provide NADPH for antioxidative effort. In addition the decrease of ascorbate and accumulation of its degradation product, threonate, indicated the activation of antioxidative pathways in menadione-treated cells. The reduced glycolytic activity probably leads to the decrease of levels of amino acids derived from glycolytic intermediates. The decrease of amino acids linked to TCA cycle intermediates and decrease of malate indicated a perturbation of TCA cycle. These observations in metabolite levels were emphasised by 13C-redistribution analysis, which indicated increased carbon flux into OPPP intermediates and inhibition of metabolic flux into all TCA cycle intermediates detected [119]. Lehmann et al. [120, 121] also conducted both metabolite profiling and 13C-redistribution analysis of menadione-treated Arabidopsis roots and found that the metabolic response of roots is distinct from that of heterotrophic cells in culture [120]. The redirection of glycolytic carbon flow and inhibition of the TCA cycle were suggested also in the roots. Especially the inhibition of the TCA cycle is more evident in roots as a perturbation of metabolite levels. In addition, roots showed pronounced accumulation of some metabolites including GABA, OAS, pyruvate, many amino acids and glucosinolates. It seems likely that cellular oxidation inhibited S assimilation and caused OAS accumulation. A general increase of amino acid levels is thought to be the result of enhanced protein degradation. This is supported by 13C-labelling analysis in which the 13C-redistribution was not affected in most amino acids, indicating that the carbon in the increased amino acids was not from synthetic pathways [121]. They also followed the metabolic recovery process after the removal of menadione from the culture media [121]. After menadione removal many of the stress-related changes reverted back to basal levels. However, each metabolic pathway recovered in a differential time period, for instance, glycolytic carbon flow reverted to control level 18 h after menadione removal, although the TCA cycle and some amino acids such as aspartate and glutamate took longer to recover. It suggests the involvement of pathway-specific regulatory processes for the oxidative stress response. These metabolic responses to menadione-induced oxidative stress mentioned above seem to be conserved among plant species and organs because quite similar responses were observed both in Arabidopsis seedlings in liquid culture [122] and rice suspension cells [123]. They are additionally at least partially similar to those observed when oxidative stress is mimicked by the removal of enzymes involved in ameliorating against it, such as manganese superoxide dismutase [124]. In the Arabidopsis plants with suppressed expression of mitochondrial manganese superoxide dismutase revealed a decrease of TCA cycle intermediates, probably because of the inhibition of aconitase and isocitrate dehydrogenase [124].
Stress combination
Whilst convenient both for experiments and discussion at the single stress level, plants are actually subjected to a combination of abiotic stress conditions in their natural habitat. Even some abiotic stresses are already combinations of stresses. For example high salt concentration causes osmotic and ion stresses, and flooding results in hypoxic and shading stresses. Although the metabolic responses of plants under a single abiotic stress have been analysed extensively as shown above, there are only few studies regarding to the effect of stress combinations on plant metabolism. Rizhsky et al. [125] applied a combination of drought and heat stress to Arabidopsis plants and analysed the metabolic profile. The metabolite profile of plants subjected to a combination of drought and heat stress was more similar to that of plants subjected to drought than to that of control plants or plants subjected to heat stress. However, the plants subjected to combined stresses accumulated high levels of sucrose and other sugars instead of proline, which is highly accumulated to a very high level in plants subjected to drought but not under stress combination. They concluded that sucrose replaces proline as the major osmoprotectant in plants subjected to combined stress because the toxic effect of high level of proline is enhanced under heat stress, as they showed experimentally [125]. Wulff-Zottele et al. [65] analysed the effect of the combination of high light irradiance and S depletion, which can occur in the field simultaneously [126]. The combination of high light and S depletion gives rise to similar metabolic pool modifications such as in high light. Proline accumulated in a differential time course under high light and stress combination. Other metabolites such as raffinose and putrescine seem to replace proline during the delay of proline accumulation in the plants subjected to high light and S depletion. This replacement of proline with sugars is similar to that observed under the combination of drought and heat stress [125]. Recently Caldana et al. [66] reported a systematic study on the metabolomic and transcriptomic response of Arabidopsis to eight environmental conditions including the combinations of changing light (darkness, high light) and/or temperature (cold and heat). The analysis has demonstrated that darkness and high temperature have a synergistic effect, thus presenting a more extreme condition. The reconstructed metabolic networks for this condition also revealed an exclusive correlation between several amino acids, including GABA with intermediates of the TCA cycle, notably succinate. These results suggested that in the absence of photosynthesis protein degradation occurs rapidly and subsequent amino acid catabolism serves as the main cellular energy supply [66].
Common and stress-specific metabolic responses against diverse abiotic stress
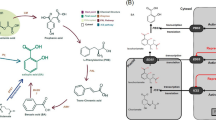
As described above, plants show a variety of metabolic responses against diverse abiotic stresses. The question is whether there are any common metabolic responses to all abiotic stresses or the responses are always specific to the stress factors. To evaluate the accumulation of these compounds under stress conditions and to search for the novel metabolic fingerprints related to the stress responses, we analysed the published metabolite profiling data available in the above-mentioned literature. Studies dealing with Arabidopsis leaves were chosen (dehydration [28], salt [79], heat and cold [57], high light and sulphur limitation [65], UV [74], light quality change [71], low nitrogen [104] and potassium limitation [104]) to afford greater comparability. Forty-five metabolites detected in more than half of the studies were analysed and each datum was converted into the fold change values against control growth conditions and presented using the log2 scale (Supplementary data, Table S1). Table 1 shows the number of metabolites accumulated or decreased under each stress condition. This reveals that abiotic stresses generally induce accumulation of metabolites with only the light quality change as an exception. The tendency of accumulation is probably related to a cessation of the growth-reducing consumption of metabolites. When subjected to abiotic stresses, plants actively re-program their growth by modulating both cell division and cell expansion. Growth decreases rapidly upon stress onset, but it recovers and adapts once stress conditions become stable [127]. Accumulated metabolites might be used as building blocks to support a recovery of growth. Figure 1 provides an overview of the changes in the amount of selected metabolites. Charts for all metabolites are found in the Supplementary data, Fig. S1. In general, changes in the amounts of metabolites were stress-specific in contrast to the general responses observed in bacteria [128]. A stress-specific change in the metabolite level would be a result of an inhibition/activation of a specific metabolic pathway especially in the short term. It should mainly be related to the properties of enzymes such as sensitivity to temperature, oxidation and ion concentration. In addition, rearrangement of the metabolic network should also result in changes of metabolites, which are related to the regulated pathways. Therefore, such a metabolite must be a good candidate for an analysis to elucidate the specific effects of an abiotic stress and the adaptive processes against it. On the other hand, metabolites responding to various stresses can be related to fundamental stress responses. In the present analysis, some metabolites can be seen to accumulate in most abiotic stress conditions although the time and extent of accumulation varied among conditions. Levels of sucrose were increased in most stress conditions in at least one time point (Fig. 1). Sucrose is a major transport sugar in most plant species and is known to accumulate under stress conditions [129]. Compounds defined as “compatible solutes” also accumulate under various abiotic stress conditions. They are very soluble in water and are non-toxic at high concentrations and function to sustain the ordered vicinal water around proteins by decreasing protein-solvent interactions at low water activities [52, 130]. This group of compounds includes betaines and related compounds; polyols and sugars, such as mannitol, sorbitol and trehalose; and amino acids, such as proline [131, 132]. Recent studies have revealed that they function to protect plants not only from osmotic stress but also from various stress factors [130, 131, 133, 134]. Therefore, the synthetic pathways of those metabolites have been of interest for metabolic engineering and some interventions have indeed increased the tolerance of some crop plants to abiotic stress [130, 131]. Raffinose is a sugar synthesised from sucrose and known to protect plant cells as an osmoprotectant; it also accumulates under most stress conditions especially at the later stages of stress treatment (Fig. 1). Raffinose is also shown to function to protect plants from oxidative damage [135], making the observation reasonable since oxidative damage likely underlies most stress conditions. Interestingly, myo-inositol, which is closely related to raffinose biosynthesis, did not show prominent changes other than under high light conditions (Fig. 1). The amino acid, proline, is known as a major compatible solute in Arabidopsis [133] and also accumulated under stress conditions despite being detected only in a limited number of studies (Fig. 1). On the other hand, trehalose accumulated only under specific conditions, suggesting that it displays functions other than being a compatible solute (Fig. 1). Indeed it is unlikely that trehalose contents in plants—other than resurrection plants—are high enough to be directly involved in stress protection [136] and some trehalose metabolism mutants exhibit potential negative effects on plant physiology [134]. The amount of trehalose may reflect that of its precursor, trehalose-6-phosphate, which has been documented to act as a signal molecule in plants [137]. GABA is another metabolite discussed in a context of stress response since it is largely and rapidly produced in response to biotic and abiotic stresses [138–140]. Our analysis supported this observation (Fig. 1). There are many suggested functions of GABA and the GABA shunt, which protect plants to survive various stress conditions including regulation of cytosolic pH, protection against oxidative stress and functions of GABA as an osmoregulator and as a signalling molecule [138]. However, whilst evidence for an important metabolic role has been documented, that for a signalling role in plants is still lacking. In the presented data set, branched chain amino acids (BCAAs), namely valine, leucine and isoleucine, and other amino acids sharing synthetic pathways with BCAA, including lysine, threonine and methionine (Fig. 2a), were generally accumulated under abiotic stress conditions. These amino acids are a novel group of metabolites that accumulated generally in response to stress conditions, although they have been shown to accumulate under drought stress conditions [141]. Joshi et al. [141] suggested that they function as compatible osmolytes since BCAA showed a high fold increase under drought stress in various plant tissues. Another possible role of BCAAs under stress conditions would be that of an alternative electron donor for the mitochondrial electron transport chain. The mitochondrial electron transport chain is primary supplied by electrons from NADH and succinate to produce ATP. Additionally there is an alternative way to feed electrons from other substrates via electron transfer flavoprotein (ETF) complex. Recent studies highlighted the importance of the alternative pathway under dark and stress conditions especially under carbon starvation (see also the "Nutrient limitation" section) [98–101]. A 13C-feeding experiment has proven that lysine and BCAA are converted into d-2-hydroxyglutarate and isovaleryl-CoA in vivo to be a direct electron donor for the ETF complex via the action of d-2-hydroxyglutarate dehydrogenase and isovaleryl-CoA dehydrogenase [101]. As such, BCAAs can provide electrons both directly to the electron transport chain via the ETF complex as well as indirectly because their catabolic products feed directly into the tricarboxylic acid (TCA) cycle [142] (Fig. 2b). The source of accumulated BCAAs would be the protein degradation product, which has recently been proposed to be an important alternative respiratory substrate especially under certain stress conditions [142], as well as the activated synthetic pathway, which is observed under drought stress conditions [28, 141]. Thus, our analysis emphasised the importance of BCAA metabolism generally under abiotic stress conditions. Interestingly, the pattern of accumulation of GABA is similar to those of BCAAs (Fig. 1), although the reason why they are related under stress conditions remains unclear. Thus, clarifying the exact mechanistic role of BCAAs under various conditions will be an important priority for the future.
Changes of the levels of metabolites in Arabidopsis leaves under various abiotic stress conditions. Each datum represents the relative metabolite level in the fold change value against control growth conditions at one time point. The values are taken from studies on dehydration [28], salt [79], heat and cold [57], high light [65], light quality change [71], UV-B light (UV) [74], low nitrogen (Low N) [104], sulphur limitation (-S) [65] and potassium limitation (-K) [116] stresses. The data set used for the analysis is found in Supplementary data, Table S1. The bars with different colours represent the values from different studies as shown in the figure. Only the metabolites of interest are shown. The charts for all metabolites are presented as Supplementary data, Fig. S1
Metabolic modes leading to the accumulation of branched chain amino acids (BCAA) and related amino acids suggested under abiotic stress conditions. a Amino acid synthetic mode. BCAAs are synthesised using pyruvate or oxaloacetate (OAA) as carbon skeletons. b Protein degradation mode. Amino acids resulting from degraded proteins would be direct and indirect electron donors to produce ATP. 2OG 2-oxoglutarate, SDH succinate dehydrogenase, HG hydroxyglutarate, D2HGDH d-2-hydroxyglutarate dehydrogenase, IVDH isovaleryl-CoA dehydrogenase, ETF electron transfer flavoprotein, ETFQO ETF-ubiquinone oxidoreductase, e − electron
Prospective: toward the elucidation of molecular mechanisms underlying abiotic stress tolerance
A wealth of metabolomics data concerning the plant stress response has been accumulated and a large number of metabolic pathways are suggested to be regulated under stress. However, there are relatively few pathways and metabolites that have been experimentally proven to function in abiotic stress tolerance. One problem is that a metabolite profile does not tell exactly whether the related metabolic pathway is up- or downregulated since both upregulation of upstream reaction and down-regulation of downstream reactions can lead to the accumulation of a metabolite. This can be solved by comparing the metabolomic data with those from transcriptomic or proteomic analysis or activities of specific enzymes [143]. Hirai et al. [111] revealed gene to metabolite regulatory networks of glucosinolate synthesis and primary metabolism under sulphur- and nitrogen-limited conditions by applying integrated analysis of transcriptome and metabolome data. Integrated analyses of the transcriptome and the metabolome successfully demonstrated connections between genes and metabolites, elucidating a wide range of signal output from ABA under dehydration [28] and the DREB1/CBF transcription factors in response to low temperature [63] as described above. This approach is proven to be useful to elucidate the regulation of the pathway and also the involvement of transcriptional regulation of the pathway. The studies using proteomics together with metabolomics are relatively rare in the plant stress response field. One example is the study by Wienkoop et al. [144], which showed the importance of starch and raffinose family oligosaccharide metabolism during temperature stress by the metabolomic and proteomic analysis of the starch-deficient Arabidopsis mutant lacking phosphoglucomutase (pgm mutant). The number of such studies should increase in the near future because of the improvement of analytical methods for proteomics. The activities of enzymes involved in a pathway should have a direct relationship with the amount of a metabolite and could be a useful tool to assess the metabolic regulation. Changes in maximal enzyme activities were analysed together with transcriptomic and metabolomic data in the study by Armengaud et al. [116], which pinpointed that pyruvate kinase activity was inhibited directly by K deficiency and was primarily responsible for the metabolic disorders observed. Metabolic flux analysis is another powerful approach to study the regulation of metabolic pathways. Lehmann et al. [121] conducted 13C redistribution analysis to prove the downregulation of glycolysis under oxidative stress treatment, which is suggested by metabolic profiling [120]. Metabolic flux analysis is also useful to elucidate a metabolic regulation that cannot be detected by metabolic profiling since the carbon flow can be affected without any apparent changes in the metabolite pool sizes [119, 145, 146]. Improvement of both experimental and theoretical mathematical approaches to flux estimation and parameterisation will greatly aid our understanding here [147]. The analysis of cell-type-specific responses would reveal more detailed mechanisms of the stress response that are hidden in a mixture of the cells in a tissue. Although the fluorescence-activated cell sorting (FACS)-based transcriptome and immunoprecipitation-based translatome data sets have provided an important foundation for the analysis of the transcriptional and translational control of environmental responses in each tissue layer of the plant [148, 149], the metabolomic studies are still rare because of the technical difficulties. Ebert et al. [150] applied single cell sampling using microcapillaries to enable the cell-type-specific metabolic analysis of epidermal cell types in Arabidopsis thaliana pavement, basal and trichome cells. Recently, Rogers et al. [151] demonstrated the feasibility of FACS-based metabolic profiling using high-resolution mass spectrometry at cell type resolution in roots.
The integration of the “omics” data revealed many molecular mechanisms for metabolic regulation but also highlighted a complex relationship among the levels of transcripts, metabolites and metabolic flux. It suggests the participation of post-transcriptional especially post-translational regulation of enzyme activity in the regulation of primary metabolism. An important role of post-transcriptional regulation in the stress response is also suggested by the poor statistical correlation of protein expression data with microarray results especially in the short-term response [152–156]. And there is an explicit indication that considerable metabolic control is executed on the metabolite and on the protein level including protein modifications [157]. Metabolic enzymes are well known to be regulated allosterically by the substrates and/or the products of the pathway [158, 159]. Many other post-translational modifications of the enzyme proteins such as phosphorylation, glutathionylation and nitrosylation could be involved in metabolic regulation [160–162]. Among them, we discuss here the reconfiguration of enzyme protein complexes in this study. In the protein complex “substrate channeling” can happen by which the intermediate produced by one enzyme is transferred to the next enzyme without complete mixing with the bulk phase [163]. Especially the association of several sequential metabolic enzymes involved in one pathway is called a metabolon [164]. Metabolite channeling can be envisioned as a means to improve catalytic efficiency by increasing local substrate concentrations, regulating competition between branch pathways for common metabolites, coordinating the activities of pathways with shared enzymes or intermediates, and sequestering reactive or toxic intermediates [165, 166]. The organisation of metabolic pathways by metabolic channeling has been discussed as the main molecular-scale organisation units to orchestrate the multiple metabolic processes and it is now supported by modelling as well as experimental evidence [167–170]. Recent bioinformatic study suggested that evolved protein interactions may contribute significantly towards increasing the efficiency of metabolic processes by permitting higher metabolic fluxes [171]. Metabolite channeling is considered to be achieved not only when the enzyme association is stable but also when the association is dynamic. Transient complexes offer the possibility of fast exchange of some of the polypeptide components upon reassembly and thus can be a molecular basis for rapid fine tuning or redirection of metabolism. The reassembly of the metabolic enzyme complex therefore should have a molecular basis underlying the metabolic regulation in mitochondria under short-term oxidative stress. Metabolic channeling including metabolon formation is reported in many processes in plants such as glycolysis, cysteine synthesis, the Calvin-Benson cycle, cyanogenic glucoside biosynthesis, the phenylpropanoid pathway, the glycine decarboxylase system and polyamine biosynthesis [165, 166]. There is however little evidence showing metabolic channeling in plant mitochondrial metabolism although the metabolon of TCA cycle enzymes and enzymes involved in amino acid metabolism are well documented in bacteria, yeast and mammals [164, 168]. More plant proteins forming enzyme super complexes should exist and contribute to metabolic regulation. Our recent study suggested that some metabolic enzymes, including malic enzyme and alanine amino transferase, altered their status in a protein complex in relation to metabolic regulation under an oxidative stress condition [122]. Further analysis is necessary to prove the involvement of enzyme protein interactions in metabolic regulation under abiotic stress conditions. Nevertheless, the expected results would lead to the new insight into plant metabolic regulation and to the full understanding of the metabolic events under abiotic stress conditions and further for the breeding of stress-tolerant crops by elucidating the target metabolic pathways to be modified.
To summarise, experiments to date have allowed us to catalogue a vast array of metabolic changes in response to stress. Without overgeneralising, since some of these are very well understood at a mechanistic level, our understanding of the causes and effects of these changes remains in some cases rather fragmentary. The metabolic changes in stress responses are considered to be divided into three phases, including a direct effect of environmental changes, transient adaptation to stress conditions and the new steady state established under prolonged stress conditions. It should be noted that each phase adopts a different duration depending on the type and the severity of the stress. A detailed time course experiment, such as those conducted in [66] or [128], is therefore necessary to distinguish to which phase the metabolic changes are related. It is also highly likely that integrating the results already obtained with those from isotope feeding experiments, comprehensive phytohormone measurements and further transcriptomic and proteomic studies will further deepen our understanding of these crucial survival processes. Once obtained such information will provide an immense foundation for metabolic engineering and synthetic biology approaches to ensuring food security.
References
Fiehn O (2001) Combining genomics, metabolome analysis, and biochemical modelling to understand metabolic networks. Comp Funct Genomics 2:155–168
Davies HV, Shepherd LV, Stewart D, Frank T, Röhlig RM, Engel KH (2010) Metabolome variability in crop plant species—when, where, how much and so what? Regul Toxicol Pharmacol 58:S54–S61
De Luca V, St Pierre B (2000) The cell and developmental biology of alkaloid biosynthesis. Trends Plant Sci 5:168–173
D’Auria JC, Gershenzon J (2005) The secondary metabolism of Arabidopsis thaliana: growing like a weed. Curr Opin Plant Biol 8:308–316
Saito K, Matsuda F (2010) Metabolomics for functional genomics, systems biology, and biotechnology. Annu Rev Plant Biol 61:463–489
Afendi FM, Okada T, Yamazaki M, Hirai-Morita A, Nakamura Y, Nakamura K, Ikeda S, Takahashi H, Altaf-Ul-Amin M, Darusman LK, Saito K, Kanaya S (2012) KNApSAcK family databases: integrated metabolite-plant species databases for multifaceted plant research. Plant Cell Physiol 53:e1
Fernie AR, Aharoni A, Willmitzer L, Stitt M, Tohge T, Kopka J, Carroll AJ, Saito K, Fraser PD, DeLuca V (2011) Recommendations for reporting metabolite data. Plant Cell 23:2477–2482
Giavalisco P, Köhl K, Hummel J, Seiwert B, Willmitzer L (2009) 13C isotope-labeled metabolomes allowing for improved compound annotation and relative quantification in liquid chromatography–mass spectrometry-based metabolomic research. Anal Chem 81:6546–6551
Fernie AR, Trethewey RN, Krotzky AJ, Willmitzer L (2004) Metabolite profiling: from diagnostics to systems biology. Nat Rev Mol Cell Bio 5:763–769
Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR (2006) Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc 1:387–396
Halket JM, Waterman D, Przyborowska AM, Patel RK, Fraser PD, Bramley PM (2005) Chemical derivatization and mass spectral libraries in metabolic profiling by GC/MS and LC/MS/MS. J Exp Bot 56:219–243
Schauer N, Steinhauser D, Strelkov S, Schomburg D, Allison G, Moritz T, Lundgren K, Roessner-Tunali U, Forbes MG, Willmitzer L, Fernie AR, Kopka J (2005) GC–MS libraries for the rapid identification of metabolites in complex biological samples. FEBS Lett 579:1332–1337
Kumari S, Stevens D, Kind T, Denkert C, Fiehn O (2011) Applying in silico retention index and mass spectra matching for identification of unknown metabolites in accurate mass GC-TOF mass spectrometry. Anal Chem 83:5895–5902
Kind T, Wohlgemuth G, Lee do Y, Lu Y, Palazoglu M, Shahbaz S, Fiehn O (2009) FiehnLib: mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal Chem 81:10038–10048
Kopka J, Schauer N, Krueger S, Birkemeyer C, Usadel B, Bergmüller E, Dörmann P, Weckwerth W, Gibon Y, Stitt M, Willmitzer L, Fernie AR, Steinhauser D (2005) GMD@CSB.DB: the Golm Metabolome Database. Bioinformatics 21:1635–1638
Rogachev I, Aharoni A (2012) UPLC-MS-based metabolite analysis in tomato. Methods Mol Biol 860:129–144
Allwood JW, Goodacre R (2010) An introduction to liquid chromatography–mass spectrometry instrumentation applied in plant metabolomic analyses. Phytochem Anal 21:33–47
Lei Z, Huhman DV, Sumner LW (2011) Mass spectrometry strategies in metabolomics. J Biol Chem 286:25435–25442
Moco S, Bino RJ, Vorst O, Verhoeven HA, de Groot J, van Beek TA, Vervoort J, de Vos CH (2006) A liquid chromatography–mass spectrometry-based metabolome database for tomato. Plant Physiol 141:1205–1218
Tohge T, Fernie AR (2009) Web-based resources for mass-spectrometry-based metabolomics: a user’s guide. Phytochemistry 70:450–456
Feldberg L, Venger I, Malitsky S, Rogachev I, Aharoni A (2009) Dual labeling of metabolites for metabolome analysis (DLEMMA): a new approach for the identification and relative quantification of metabolites by means of dual isotope labeling and liquid chromatography–mass spectrometry. Anal Chem 81:9257–9266
Arrivault S, Guenther M, Ivakov A, Feil R, Vosloh D, van Dongen JT, Sulpice R, Stitt M (2009) Use of reverse-phase liquid chromatography, linked to tandem mass spectrometry, to profile the Calvin cycle and other metabolic intermediates in Arabidopsis rosettes at different carbon dioxide concentrations. Plant J 59:826–839
Kanno Y, Jikumaru Y, Hanada A, Nambara E, Abrams SR, Kamiya Y, Seo M (2010) Comprehensive hormone profiling in developing Arabidopsis seeds: examination of the site of ABA biosynthesis, ABA transport and hormone interactions. Plant Cell Physiol 51:1988–2001
Ramautar R, Somsen GW, de Jong GJ (2009) CE-MS in metabolomics. Electrophoresis 30:276–291
Ramautar R, Mayboroda OA, Somsen GW, de Jong GJ (2011) CE-MS for metabolomics: developments and applications in the period 2008–2010. Electrophoresis 32:52–65
Monton MR, Soga T (2007) Metabolome analysis by capillary electrophoresis–mass spectrometry. J Chromatogr A 1168:237–246
Sugimoto M, Hirayama A, Robert M, Abe S, Soga T, Tomita M (2010) Prediction of metabolite identity from accurate mass, migration time prediction and isotopic pattern information in CE-TOFMS data. Electrophoresis 31:2311–2318
Urano K, Maruyama K, Ogata Y, Morishita Y, Takeda M, Sakurai N, Suzuki H, Saito K, Shibata D, Kobayashi M, Yamaguchi-Shinozaki K, Shinozaki K (2009) Characterization of the ABA-regulated global responses to dehydration in Arabidopsis by metabolomics. Plant J 57:1065–1078
Williams BJ, Cameron CJ, Workman R, Broeckling CD, Sumner LW, Smith JT (2007) Amino acid profiling in plant cell cultures: an inter-laboratory comparison of CE-MS and GC–MS. Electrophoresis 28:1371–1379
Soga T, Imaizumi M (2001) Capillary electrophoresis method for the analysis of inorganic anions, organic acids, amino acids, nucleotides, carbohydrates and other anionic compounds. Electrophoresis 22:3418–3425
Fernie AR (2007) The future of metabolic phytochemistry: larger numbers of metabolites, higher resolution, greater understanding. Phytochemistry 68:2861–2880
Schripsema J (2010) Application of NMR in plant metabolomics: techniques, problems and prospects. Phytochem Anal 21:14–21
Terskikh VV, Feurtado JA, Borchardt S, Giblin M, Abrams SR, Kermode AR (2005) In vivo 13C NMR metabolite profiling: potential for understanding and assessing conifer seed quality. J Exp Bot 56:2253–2265
Ratcliffe RG, Shachar-Hill Y (2005) Revealing metabolic phenotypes in plants: inputs from NMR analysis. Biol Rev Camb Philos Soc 80:27–43
Eisenreich W, Bacher A (2007) Advances of high-resolution NMR techniques in the structural and metabolic analysis of plant biochemistry. Phytochemistry 68:2799–2815
Krishnan P, Kruger NJ, Ratcliffe RG (2005) Metabolite fingerprinting and profiling in plants using NMR. J Exp Bot 56:255–265
Martin FP, Dumas ME, Wang Y, Legido-Quigley C, Yap IK, Tang H, Zirah S, Murphy GM, Cloarec O, Lindon JC, Sprenger N, Fay LB, Kochhar S, van Bladeren P, Holmes E, Nicholson JK (2007) A top-down systems biology view of microbiome-mammalian metabolic interactions in a mouse model. Mol Syst Biol 3:112
Lommen A, Weseman JM, Smith GO, Noteborn HPJM (1998) On the detection of environmental effects on complex matrices combining off-line liquid chromatography and 1H-NMR. Biodegradation 9:513–525
Dixon RA, Gang DR, Charlton AJ, Fiehn O, Kuiper HA, Reynolds TL, Tjeerdema RS, Jeffery EH, German JB, Ridley WP, Seiber JN (2006) Applications of metabolomics in agriculture. J Agric Food Chem 54:8984–8994
Shulaev V, Cortes D, Miller G, Mittler R (2008) Metabolomics for plant stress response. Physiol Plant 132:199–208
Bowne J, Bacic A, Tester MRU (2011) Abiotic stress and metabolomics. Annu Plant Rev 43:61–85
Cominelli E, Galbiati M, Tonelli C, Bowler C (2009) Water: the invisible problem. Access to fresh water is considered to be a universal and free human right, but dwindling resources and a burgeoning population are increasing its economic value. EMBO Rep 10:671–676
Chaves MM, Oliveira MM (2004) Mechanisms underlying plant resilience to water deficits: prospects for water-saving agriculture. J Exp Bot 55:2365–2384
Verslues PE, Juenger TE (2011) Drought, metabolites, and Arabidopsis natural variation: a promising combination for understanding adaptation to water-limited environments. Curr Opin Plant Biol 14:240–245
Skirycz A, Vandenbroucke K, Clauw P, Maleux K, De Meyer B, Dhondt S, Pucci A, Gonzalez N, Hoeberichts F, Tognetti VB, Galbiati M, Tonelli C, Van Breusegem F, Vuylsteke M, Inzé D (2011) Survival and growth of Arabidopsis plants given limited water are not equal. Nat Biotechnol 29:212–214
Skirycz A, De Bodt S, Obata T, De Clercq I, Claeys H, De Rycke R, Andriankaja M, Van Aken O, Van Breusegem F, Fernie AR, Inzé D (2010) Developmental stage specificity and the role of mitochondrial metabolism in the response of Arabidopsis leaves to prolonged mild osmotic stress. Plant Physiol 152:226–244
Semel Y, Nissenbaum J, Menda N, Zinder M, Krieger U, Issman N, Pleban T, Lippman Z, Gur A, Zamir D (2006) Overdominant quantitative trait loci for yield and fitness in tomato. Proc Natl Acad Sci USA 103:12981–12986
Witt S, Galicia L, Lisec J, Cairns J, Tiessen A, Araus JL, Palacios-Rojas N, Fernie AR (2012) Metabolic and phenotypic responses of greenhouse-grown maize hybrids to experimentally controlled drought stress. Mol Plant 5:401–417
Bowne JB, Erwin TA, Juttner J, Schnurbusch T, Langridge P, Bacic A, Roessner U (2012) Drought responses of leaf tissues from wheat cultivars of differing drought tolerance at the metabolite level. Mol Plant 5:418–429
van Dongen JT, Fröhlich A, Ramírez-Aguilar SJ, Schauer N, Fernie AR, Erban A, Kopka J, Clark J, Langer A, Geigenberger P (2009) Transcript and metabolite profiling of the adaptive response to mild decreases in oxygen concentration in the roots of Arabidopsis plants. Ann Bot 103:269–280
Rocha M, Licausi F, Araújo WL, Nunes-Nesi A, Sodek L, Fernie AR, van Dongen JT (2010) Glycolysis and the tricarboxylic acid cycle are linked by alanine aminotransferase during hypoxia induced by waterlogging of Lotus japonicus. Plant Physiol 152:1501–1513
Guy CL (1990) Cold Acclimation and Freezing Stress Tolerance: Role of Protein Metabolism. Annu Rev Plant Biol Plant Mol Biol 41:187–223
Wanner LA, Junttila O (1999) Cold-induced freezing tolerance in Arabidopsis. Plant Physiol 120:391–400
Thomashow MF (2010) Molecular basis of plant cold acclimation: insights gained from studying the CBF cold response pathway. Plant Physiol 154:571–577
Medina J, Catalá R, Salinas J (2011) The CBFs: three Arabidopsis transcription factors to cold acclimate. Plant Sci 180:3–11
Cook D, Fowler S, Fiehn O, Thomashow MF (2004) A prominent role for the CBF cold response pathway in configuring the low-temperature metabolome of Arabidopsis. Proc Natl Acad Sci USA 101:15243–15248
Kaplan F, Kopka J, Haskell DW, Zhao W, Schiller KC, Gatzke N, Sung DY, Guy CL (2004) Exploring the temperature-stress metabolome. Plant Physiol 136:4159–4168
Kaplan F, Kopka J, Sung DY, Zhao W, Popp M, Porat R, Guy CL (2007) Transcript and metabolite profiling during cold acclimation of Arabidopsis reveals an intricate relationship of cold-regulated gene expression with modifications in metabolite content. Plant J 50:967–981
Gray GR, Heath D (2005) A global reorganization of the metabolome in Arabidopsis during cold acclimation is revealed by metabolic fingerprinting. Physiol Plant 124:236–248
Hannah MA, Wiese D, Freund S, Fiehn O, Heyer AG, Hincha DK (2006) Natural genetic variation of freezing tolerance in Arabidopsis. Plant Physiol 142:98–112
Espinoza C, Degenkolbe T, Caldana C, Zuther E, Leisse A, Willmitzer L, Hincha DK, Hannah MA (2010) Interaction with diurnal and circadian regulation results in dynamic metabolic and transcriptional changes during cold acclimation in Arabidopsis. PLoS One 5:e14101
Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10:1391–1406
Maruyama K, Takeda M, Kidokoro S, Yamada K, Sakuma Y, Urano K, Fujita M, Yoshiwara K, Matsukura S, Morishita Y, Sasaki R, Suzuki H, Saito K, Shibata D, Shinozaki K, Yamaguchi-Shinozaki K (2009) Metabolic pathways involved in cold acclimation identified by integrated analysis of metabolites and transcripts regulated by DREB1A and DREB2A. Plant Physiol 150:1972–1980
Lugan R, Niogret MF, Kervazo L, Larher FR, Kopka J, Bouchereau A (2009) Metabolome and water status phenotyping of Arabidopsis under abiotic stress cues reveals new insight into ESK1 function. Plant Cell Environ 32:95–108
Wulff-Zottele C, Gatzke N, Kopka J, Orellana A, Hoefgen R, Fisahn J, Hesse H (2010) Photosynthesis and metabolism interact during acclimation of Arabidopsis thaliana to high irradiance and sulphur depletion. Plant Cell Environ 33:1974–1988
Caldana C, Degenkolbe T, Cuadros-Inostroza A, Klie S, Sulpice R, Leisse A, Steinhauser D, Fernie AR, Willmitzer L, Hannah MA (2011) High-density kinetic analysis of the metabolomic and transcriptomic response of Arabidopsis to eight environmental conditions. Plant J 67:869–884
Terashima I, Araya T, Miyazawa S, Sone K, Yano S (2005) Construction and maintenance of the optimal photosynthetic systems of the leaf, herbaceous plant and tree: an eco-developmental treatise. Ann Bot 95:507–519
Smith H (2000) Phytochromes and light signal perception by plants—an emerging synthesis. Nature 407:585–591
Allen JF (2003) Cyclic, pseudocyclic and noncyclic photophosphorylation: new links in the chain. Trends Plant Sci 8:15–19
Dietzel L, Bräutigam K, Pfannschmidt T (2008) Photosynthetic acclimation: state transitions and adjustment of photosystem stoichiometry–functional relationships between short-term and long-term light quality acclimation in plants. FEBS J 275:1080–1088
Bräutigam K, Dietzel L, Kleine T, Ströher E, Wormuth D, Dietz KJ, Radke D, Wirtz M, Hell R, Dörmann P, Nunes-Nesi A, Schauer N, Fernie AR, Oliver SN, Geigenberger P, Leister D, Pfannschmidt T (2009) Dynamic plastid redox signals integrate gene expression and metabolism to induce distinct metabolic states in photosynthetic acclimation in Arabidopsis. Plant Cell 21:2715–2732
Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C (2000) Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J 19:6150–6161
Ries G, Heller W, Puchta H, Sandermann H, Seidlitz HK, Hohn B (2000) Elevated UV-B radiation reduces genome stability in plants. Nature 406:98–101
Kusano M, Tohge T, Fukushima A, Kobayashi M, Hayashi N, Otsuki H, Kondou Y, Goto H, Kawashima M, Matsuda F, Niida R, Matsui M, Saito K, Fernie AR (2011) Metabolomics reveals comprehensive reprogramming involving two independent metabolic responses of Arabidopsis to UV-B light. Plant J 67:354–369
Tohge T, Kusano M, Fukushima A, Saito K, Fernie AR (2011) Transcriptional and metabolic programs following exposure of plants to UV-B irradiation. Plant Signal Behav 6:1987–1992
Munns R (2005) Genes and salt tolerance: bringing them together. New Phytol 167:645–663
Kim JK, Bamba T, Harada K, Fukusaki E, Kobayashi A (2007) Time-course metabolic profiling in Arabidopsis thaliana cell cultures after salt stress treatment. J Exp Bot 58:415–424
Tester M, Davenport R (2003) Na+ tolerance and Na+ transport in higher plants. Ann Bot 91:503–527
Gong Q, Li P, Ma S, Indu Rupassara S, Bohnert HJ (2005) Salinity stress adaptation competence in the extremophile Thellungiella halophila in comparison with its relative Arabidopsis thaliana. Plant J 44:826–839
Lugan R, Niogret MF, Leport L, Guégan JP, Larher FR, Savouré A, Kopka J, Bouchereau A (2010) Metabolome and water homeostasis analysis of Thellungiella salsuginea suggests that dehydration tolerance is a key response to osmotic stress in this halophyte. Plant J 64:215–229
Johnson HE, Broadhurst D, Goodacre R, Smith AR (2003) Metabolic fingerprinting of salt-stressed tomatoes. Phytochemistry 62:919–928
Cramer GR, Ergül A, Grimplet J, Tillett RL, Tattersall EA, Bohlman MC, Vincent D, Sonderegger J, Evans J, Osborne C, Quilici D, Schlauch KA, Schooley DA, Cushman JC (2007) Water and salinity stress in grapevines: early and late changes in transcript and metabolite profiles. Funct Integr Genomics 7:111–134
Brosché M, Vinocur B, Alatalo ER, Lamminmäki A, Teichmann T, Ottow EA, Djilianov D, Afif D, Bogeat-Triboulot MB, Altman A, Polle A, Dreyer E, Rudd S, Paulin L, Auvinen P, Kangasjärvi J (2005) Gene expression and metabolite profiling of Populus euphratica growing in the Negev desert. Genome Biol 6:R101
Gagneul D, Aïnouche A, Duhazé C, Lugan R, Larher FR, Bouchereau A (2007) A reassessment of the function of the so-called compatible solutes in the halophytic plumbaginaceae Limonium latifolium. Plant Physiol 144:1598–1611
Zuther E, Koehl K, Kopka J (2007) Comparative metabolome analysis of the salt response in breeding cultivars of rice. In: Jenks MA, Hasegawa PM, Jain SM (eds) Advances in molecular breeding toward drought and salt tolerant crops. Springer, Berlin, pp 285–315
Sanchez DH, Siahpoosh MR, Roessner U, Udvardi M, Kopka J (2008) Plant metabolomics reveals conserved and divergent metabolic responses to salinity. Physiol Plant 132:209–219
Sanchez DH, Lippold F, Redestig H, Hannah MA, Erban A, Krämer U, Kopka J, Udvardi MK (2008) Integrative functional genomics of salt acclimatization in the model legume Lotus japonicus. Plant J 53:973–987
Sanchez DH, Pieckenstain FL, Escaray F, Erban A, Kraemer U, Udvardi MK, Kopka J (2011) Comparative ionomics and metabolomics in extremophile and glycophytic Lotus species under salt stress challenge the metabolic pre-adaptation hypothesis. Plant Cell Environ 34:605–617
Sanchez DH, Pieckenstain FL, Szymanski J, Erban A, Bromke M, Hannah MA, Kraemer U, Kopka J, Udvardi MK (2011) Comparative functional genomics of salt stress in related model and cultivated plants identifies and overcomes limitations to translational genomics. PLoS One 6:e17094
Sharma SS, Dietz KJ (2009) The relationship between metal toxicity and cellular redox imbalance. Trends Plant Sci 14:43–50
Jahangir M, Abdel-Farid IB, Choi YH, Verpoorte R (2008) Metal ion-inducing metabolite accumulation in Brassica rapa. J Plant Physiol 165:1429–1437
Sun X, Zhang J, Zhang H, Ni Y, Zhang Q, Chen J, Guan Y (2010) The responses of Arabidopsis thaliana to cadmium exposure explored via metabolite profiling. Chemosphere 78:840–845
Dubey S, Misra P, Dwivedi S, Chatterjee S, Bag SK, Mantri S, Asif MH, Rai A, Kumar S, Shri M, Tripathi P, Tripathi RD, Trivedi PK, Chakrabarty D, Tuli R (2010) Transcriptomic and metabolomic shifts in rice roots in response to Cr(VI) stress. BMC Genomics 11:648
Smith AM, Stitt M (2007) Coordination of carbon supply and plant growth. Plant Cell Environ 30:1126–1149
Stitt M, Gibon Y, Lunn JE, Piques M (2007) Multilevel genomics analysis of carbon signalling during low carbon availability: coordinating the supply and utilisation of carbon in a fluctuating environment. Funct Plant Biol 34:526
Osuna D, Usadel B, Morcuende R, Gibon Y, Bläsing OE, Höhne M, Günter M, Kamlage B, Trethewey R, Scheible WR, Stitt M (2007) Temporal responses of transcripts, enzyme activities and metabolites after adding sucrose to carbon-deprived Arabidopsis seedlings. Plant J 49:463–491
Usadel B, Bläsing OE, Gibon Y, Retzlaff K, Höhne M, Günther M, Stitt M (2008) Global transcript levels respond to small changes of the carbon status during progressive exhaustion of carbohydrates in Arabidopsis rosettes. Plant Physiol 146:1834–1861
Ishizaki K, Larson TR, Schauer N, Fernie AR, Graham IA, Leaver CJ (2005) The critical role of Arabidopsis electron-transfer flavoprotein:ubiquinone oxidoreductase during dark-induced starvation. Plant Cell 17:2587–2600
Ishizaki K, Larson TR, Schauer N, Fernie AR, Graham IA, Leaver CJ (2006) The mitochondrial electron transfer flavoprotein complex is essential for survival of Arabidopsis in extended darkness. Plant J 47:751–760
Araújo WL, Ishizaki K, Nunes-Nesi A, Tohge T, Larson TR, Krahnert I, Balbo I, Witt S, Dörmann P, Graham IA, Leaver CJ, Fernie AR (2011) Analysis of a range of catabolic mutants provides evidence that phytanoyl-coenzyme A does not act as a substrate of the electron-transfer flavoprotein/electron-transfer flavoprotein:ubiquinone oxidoreductase complex in Arabidopsis during dark-induced sene. Plant Physiol 157:55–69
Araújo WL, Ishizaki K, Nunes-Nesi A, Larson TR, Tohge T, Krahnert I, Witt S, Obata T, Schauer N, Graham IA, Leaver CJ, Fernie AR (2010) Identification of the 2-hydroxyglutarate and isovaleryl-CoA dehydrogenases as alternative electron donors linking lysine catabolism to the electron transport chain of Arabidopsis mitochondria. Plant Cell 22:1549–1563
Urbanczyk-Wochniak E, Fernie AR (2005) Metabolic profiling reveals altered nitrogen nutrient regimes have diverse effects on the metabolism of hydroponically-grown tomato (Solanum lycopersicum) plants. J Exp Bot 56:309–321
Stitt M, Fernie AR (2003) From measurements of metabolites to metabolomics: an “on the fly” perspective illustrated by recent studies of carbon–nitrogen interactions. Curr Opin Biotechnol 14:136–144
Tschoep H, Gibon Y, Carillo P, Armengaud P, Szecowka M, Nunes-Nesi A, Fernie AR, Koehl K, Stitt M (2009) Adjustment of growth and central metabolism to a mild but sustained nitrogen-limitation in Arabidopsis. Plant Cell Environ 32:300–318
Morcuende R, Bari R, Gibon Y, Zheng W, Pant BD, Bläsing O, Usadel B, Czechowski T, Udvardi MK, Stitt M, Scheible WR (2007) Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant Cell Environ 30:85–112
Hernández G, Ramírez M, Valdés-López O, Tesfaye M, Graham MA, Czechowski T, Schlereth A, Wandrey M, Erban A, Cheung F, Wu HC, Lara M, Town CD, Kopka J, Udvardi MK, Vance CP (2007) Phosphorus stress in common bean: root transcript and metabolic responses. Plant Physiol 144:752–767
Hernández G, Valdés-López O, Ramírez M, Goffard N, Weiller G, Aparicio-Fabre R, Fuentes SI, Erban A, Kopka J, Udvardi MK, Vance CP (2009) Global changes in the transcript and metabolic profiles during symbiotic nitrogen fixation in phosphorus-stressed common bean plants. Plant Physiol 151:1221–1238
Huang CY, Roessner U, Eickmeier I, Genc Y, Callahan DL, Shirley N, Langridge P, Bacic A (2008) Metabolite profiling reveals distinct changes in carbon and nitrogen metabolism in phosphate-deficient barley plants (Hordeum vulgare L.). Plant Cell Physiol 49:691–703
Nikiforova VJ, Daub CO, Hesse H, Willmitzer L, Hoefgen R (2005) Integrative gene-metabolite network with implemented causality deciphers informational fluxes of sulphur stress response. J Exp Bot 56:1887–1896
Nikiforova VJ, Kopka J, Tolstikov V, Fiehn O, Hopkins L, Hawkesford MJ, Hesse H, Hoefgen R (2005) Systems rebalancing of metabolism in response to sulfur deprivation, as revealed by metabolome analysis of Arabidopsis plants. Plant Physiol 138:304–318
Hirai MY, Yano M, Goodenowe DB, Kanaya S, Kimura T, Awazuhara M, Arita M, Fujiwara T, Saito K (2004) Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proc Natl Acad Sci USA 101:10205–10210
Hirai MY, Klein M, Fujikawa Y, Yano M, Goodenowe DB, Yamazaki Y, Kanaya S, Nakamura Y, Kitayama M, Suzuki H, Sakurai N, Shibata D, Tokuhisa J, Reichelt M, Gershenzon J, Papenbrock J, Saito K (2005) Elucidation of gene-to-gene and metabolite-to-gene networks in Arabidopsis by integration of metabolomics and transcriptomics. J Biol Chem 280:25590–25595
Hoefgen R, Nikiforova VJ (2008) Metabolomics integrated with transcriptomics: assessing systems response to sulfur-deficiency stress. Physiol Plant 132:190–198
Hubberten HM, Klie S, Caldana C, Degenkolbe T, Willmitzer L, Hoefgen R (2012) An additional role of O-acetylserine as a sulphur status independent regulator during plant growth. Plant J 70:666–677
Marschner H (1995) Mineral nutrition of higher plants, 2nd edn. Academic Press, London
Armengaud P, Sulpice R, Miller AJ, Stitt M, Amtmann A, Gibon Y (2009) Multilevel analysis of primary metabolism provides new insights into the role of potassium nutrition for glycolysis and nitrogen assimilation in Arabidopsis roots. Plant Physiol 150:772–785
Mittler R (2006) Abiotic stress, the field environment and stress combination. Trends Plant Sci 11:15–19
Møller IM, Jensen PE, Hansson A (2007) Oxidative modifications to cellular components in plants. Annu Rev Plant Biol 58:459–481
Baxter CJ, Redestig H, Schauer N, Repsilber D, Patil KR, Nielsen J, Selbig J, Liu J, Fernie AR, Sweetlove LJ (2007) The metabolic response of heterotrophic Arabidopsis cells to oxidative stress. Plant Physiol 143:312–325
Lehmann M, Schwarzländer M, Obata T, Sirikantaramas S, Burow M, Olsen CE, Tohge T, Fricker MD, Møller BL, Fernie AR, Sweetlove LJ, Laxa M (2009) The metabolic response of Arabidopsis roots to oxidative stress is distinct from that of heterotrophic cells in culture and highlights a complex relationship between the levels of transcripts, metabolites, and flux. Mol Plant 2:390–406
Lehmann M, Laxa M, Sweetlove LJ, Fernie AR, Obata T (2011) Metabolic recovery of Arabidopsis thaliana roots following cessation of oxidative stress. Metabolomics 8:143–153
Obata T, Matthes A, Koszior S, Lehmann M, Araújo WL, Bock R, Sweetlove LJ, Fernie AR (2011) Alteration of mitochondrial protein complexes in relation to metabolic regulation under short-term oxidative stress in Arabidopsis seedlings. Phytochemistry 72:1081–1091
Ishikawa T, Takahara K, Hirabayashi T, Matsumura H, Fujisawa S, Terauchi R, Uchimiya H, Kawai-Yamada M (2010) Metabolome analysis of response to oxidative stress in rice suspension cells overexpressing cell death suppressor Bax inhibitor-1. Plant Cell Physiol 51:9–20
Morgan MJ, Lehmann M, Schwarzländer M, Baxter CJ, Sienkiewicz-Porzucek A, Williams TC, Schauer N, Fernie AR, Fricker MD, Ratcliffe RG, Sweetlove LJ, Finkemeier I (2008) Decrease in manganese superoxide dismutase leads to reduced root growth and affects tricarboxylic acid cycle flux and mitochondrial redox homeostasis. Plant Physiol 147:101–114
Rizhsky L, Liang H, Shuman J, Shulaev V, Davletova S, Mittler R (2004) When defense pathways collide. The response of Arabidopsis to a combination of drought and heat stress. Plant Physiol 134:1683–1696
Buri M, Masunaga T, Wakatsuki T (2000) Sulfur and zinc levels as limiting factors to rice production in West Africa lowlands. Geoderma 94:23–42
Skirycz A, Inzé D (2010) More from less: plant growth under limited water. Curr Opin Biotechnol 21:197–203
Jozefczuk S, Klie S, Catchpole G, Szymanski J, Cuadros-Inostroza A, Steinhauser D, Selbig J, Willmitzer L (2010) Metabolomic and transcriptomic stress response of Escherichia coli. Mol Syst Biol 6:364
Rolland F, Baena-Gonzalez E, Sheen J (2006) Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu Rev Plant Biol 57:675–709
Chen THH, Murata N (2011) Glycinebetaine protects plants against abiotic stress: mechanisms and biotechnological applications. Plant Cell Environ 34:1–20
Chen THH, Murata N (2002) Enhancement of tolerance of abiotic stress by metabolic engineering of betaines and other compatible solutes. Curr Opin Plant Biol 5:250–257
Diamant S, Eliahu N, Rosenthal D, Goloubinoff P (2001) Chemical chaperones regulate molecular chaperones in vitro and in cells under combined salt and heat stresses. J Biol Chem 276:39586–395891
Verbruggen N, Hermans C (2008) Proline accumulation in plants: a review. Amino Acids 35:753–759
Fernandez O, Béthencourt L, Quero A, Sangwan RS, Clément C (2010) Trehalose and plant stress responses: friend or foe? Trends Plant Sci 15:409–417
Nishizawa A, Yabuta Y, Shigeoka S (2008) Galactinol and raffinose constitute a novel function to protect plants from oxidative damage. Plant Physiol 147:1251–1263
Wingler A (2002) The function of trehalose biosynthesis in plants. Phytochemistry 60:437–440
Smeekens S, Ma J, Hanson J, Rolland F (2010) Sugar signals and molecular networks controlling plant growth. Curr Opin Plant Biol 13:274–279
Bouché N, Fromm H (2004) GABA in plants: just a metabolite? Trends Plant Sci 9:110–115
Shelp BJ, Bown AW, Mclean MD (1999) Metabolism and functions of gamma-aminobutyric acid. Trends Plant Sci 4:446–452
Kinnersley A, Turano F (2000) Gamma aminobutyric acid (GABA) and plant responses to stress. Crit Rev Plant Sci 19:479–509
Joshi V, Joung JG, Fei Z, Jander G (2010) Interdependence of threonine, methionine and isoleucine metabolism in plants: accumulation and transcriptional regulation under abiotic stress. Amino Acids 39:933–947
Araújo WL, Tohge T, Ishizaki K, Leaver CJ, Fernie AR (2011) Protein degradation—an alternative respiratory substrate for stressed plants. Trends Plant Sci 16:489–498
Cramer GR, Urano K, Delrot S, Pezzotti M, Shinozaki K (2011) Effects of abiotic stress on plants: a systems biology perspective. BMC Plant Biol 11:163
Wienkoop S, Morgenthal K, Wolschin F, Scholz M, Selbig J, Weckwerth W (2008) Integration of metabolomic and proteomic phenotypes: analysis of data covariance dissects starch and RFO metabolism from low and high temperature compensation response in Arabidopsis thaliana. Mol Cell Proteomics 7:1725–1736
Fell DA (2005) Enzymes, metabolites and fluxes. J Exp Bot 56:267–272
Fernie AR, Geigenberger P, Stitt M (2005) Flux an important, but neglected, component of functional genomics. Curr Opin Plant Biol 8:174–182
Fernie AR, Stitt M (2012) On the discordance of metabolomics with proteomics and transcriptomics: coping with increasing complexity in logic, chemistry and network interactions. Plant Physiol 158:1139–1145
Dinneny JR, Long TA, Wang JY, Jung JW, Mace D, Pointer S, Barron C, Brady SM, Schiefelbein J, Benfey PN (2008) Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science 320:942–945
Wee CW, Dinneny JR (2010) Tools for high-spatial and temporal-resolution analysis of environmental responses in plants. Biotechnol Lett 32:1361–1371
Ebert B, Zöller D, Erban A, Fehrle I, Hartmann J, Niehl A, Kopka J, Fisahn J (2010) Metabolic profiling of Arabidopsis thaliana epidermal cells. J Exp Bot 61:1321–1335
Rogers ED, Jackson T, Moussaieff A, Aharoni A, Benfey PN (2012) Cell type-specific transcriptional profiling: implications for metabolite profiling. Plant J 70:5–17
Koussevitzky S, Suzuki N, Huntington S, Armijo L, Sha W, Cortes D, Shulaev V, Mittler R (2008) Ascorbate peroxidase 1 plays a key role in the response of Arabidopsis thaliana to stress combination. J Biol Chem 283:34197–34203
Bogeat-Triboulot MB, Brosché M, Renaut J, Jouve L, Le Thiec D, Fayyaz P, Vinocur B, Witters E, Laukens K, Teichmann T, Altman A, Hausman JF, Polle A, Kangasjärvi J, Dreyer E (2007) Gradual soil water depletion results in reversible changes of gene expression, protein profiles, ecophysiology, and growth performance in Populus euphratica, a poplar growing in arid regions. Plant Physiol 143:876–892
Grimplet J, Wheatley MD, Jouira HB, Deluc LG, Cramer GR, Cushman JC (2009) Proteomic and selected metabolite analysis of grape berry tissues under well-watered and water-deficit stress conditions. Proteomics 9:2503–2528
Nie L, Wu G, Culley DE, Scholten JC, Zhang W (2007) Integrative analysis of transcriptomic and proteomic data: challenges, solutions and applications. Crit Rev Biotechnol 27:63–75
Böhmer M, Schroeder JI (2011) Quantitative transcriptomic analysis of abscisic acid-induced and reactive oxygen species-dependent expression changes and proteomic profiling in Arabidopsis suspension cells. Plant J 67:105–118
Morgenthal K, Wienkoop S, Wolschin F, Weckwerth W (2007) Integrative profiling of metabolites and proteins: improving pattern recognition and biomarker selection for systems level approaches. Methods Mol Biol 358:57–75
Huang Z, Zhu L, Cao Y, Wu G, Liu X, Chen Y, Wang Q, Shi T, Zhao Y, Wang Y, Li W, Li Y, Chen H, Chen G, Zhang J (2011) ASD: a comprehensive database of allosteric proteins and modulators. Nucleic Acids Res 39:D663–D669
Goodey NM, Benkovic SJ (2008) Allosteric regulation and catalysis emerge via a common route. Nat Chem Biol 4:474–482
Dalle-Donne I, Rossi R, Giustarini D, Colombo R, Milzani A (2007) S-glutathionylation in protein redox regulation. Free Radic Biol Med 43:883–898
Lindermayr C, Durner J (2009) S-Nitrosylation in plants: pattern and function. J Proteomics 73:1–9
Schulze WX (2010) Proteomics approaches to understand protein phosphorylation in pathway modulation. Curr Opin Plant Biol 13:280–287
Spivey HO, Ovádi J (1999) Substrate channeling. Methods 19:306–321
Srere PA (1987) Complexes of sequential metabolic enzymes. Annu Rev Biochem 56:89–124
Winkel BSJ (2004) Metabolic channeling in plants. Annu Rev Plant Biol 55:85–107
Jørgensen K, Rasmussen AV, Morant M, Nielsen AH, Bjarnholt N, Zagrobelny M, Bak S, Møller BL (2005) Metabolon formation and metabolic channeling in the biosynthesis of plant natural products. Curr Opin Plant Biol 8:280–291
Graham JW, Williams TC, Morgan M, Fernie AR, Ratcliffe RG, Sweetlove LJ (2007) Glycolytic enzymes associate dynamically with mitochondria in response to respiratory demand and support substrate channeling. Plant Cell 19:3723–3738
Srere PA (2000) Macromolecular interactions: tracing the roots. Trends Biochem Sci 25:150–153
Degenring D, Röhl M, Uhrmacher AM (2004) Discrete event, multi-level simulation of metabolite channeling. Biosystems 75:29–41
Ro DK, Douglas CJ (2004) Reconstitution of the entry point of plant phenylpropanoid metabolism in yeast (Saccharomyces cerevisiae): implications for control of metabolic flux into the phenylpropanoid pathway. J Biol Chem 279:2600–2607
Durek P, Walther D (2008) The integrated analysis of metabolic and protein interaction networks reveals novel molecular organizing principles. BMC Syst Biol 2:100
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Obata, T., Fernie, A.R. The use of metabolomics to dissect plant responses to abiotic stresses. Cell. Mol. Life Sci. 69, 3225–3243 (2012). https://doi.org/10.1007/s00018-012-1091-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-012-1091-5