Abstract
Beginning in the first decade of 1900, pioneering research in disease resistance and seed color inheritance established the scientific basis of Mendelian inheritance in wheat breeding. A series of breakthroughs in chromosome and genome analysis beginning in the 1920s and continuing into the twenty-first century have impacted wheat improvement. The application of meiotic chromosome pairing in the 1920s and plasmon analysis in the 1950s elucidated phylogeny of the Triticum-Aegilops complex of species and defined the wheat gene pools. The aneuploid stocks in the 1950s opened floodgates for chromosome and arm mapping of first phenotypic and later protein and DNA probes. The aneuploid stocks, coupled with advances in chromosome banding and in situ hybridization in the 1970s, allowed precise chromosome engineering of traits in wide hybrids. The deletion stocks in the 1990s were pivotal in mapping expressed genes to specific chromosome bins revealing structural and functional differentiation of chromosomes along their length and facilitating map-based cloning of genes. Advances in whole-genome sequencing, chromosome genomics, RH mapping and functional tools led to the assembly of reference sequence of Chinese Spring and multiple wheat genomes. Chromosome and genomic analysis must be integrated into wheat breeding and wide-hybridizaton pipeline for sustainable crop improvement.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
- Genome analyzer methods
- Wheat phylogeny
- Aneuploidy
- Chromosome banding
- in situ hybridization
- Deletion stocks
- Genome sequencing
1 Learning Objectives
-
Become familiar with the history of wheat genetics, cytogenetics and genomics research, the scientists who did the work, the significance of their discoveries and how it impacted wheat genetics and breeding research.
2 Introduction
The author [1] had the pleasure of doing graduate work with Professor Charley Rick, who obtained his PhD with Karl Sax (pioneer wheat cytogeneticist) at Harvard University in 1940 and the same year began his career at UC Davis. I did postdoctoral research (1973–1975) with Dr. Ernie Sears (Father of Wheat Genetics), who obtained his PhD with EM East, Harvard University in 1936, and the same year began his research career with USDA at the University of Missouri. My co-supervisor at the University of Missouri was Professor Gordon Kimber, who trained at the famous Plant Breeding Institute at Cambridge in UK. As a founding director of Wheat Genetics Resource Center (1984–present), a position from which I retired in 2018, we conducted collaborative research with major wheat research groups in the US and worldwide, including CIMMYT and ICARDA [2]. Many of my academic pedigree and first and second generation scientists are active in crop research. From this vantage point, I want to highlight major breakthroughs over a century of wheat cytogenetic and genome analysis research and how it impacted crop improvement. Due to limitations on space and citations, for original citations, the reader may be referred to secondary citations in books [3,4,5,6] or review articles [7,8,9,10,11].
3 Validation of Mendel’s Laws of Inheritance in Wheat Laid the Foundation for Scientific Breeding
Soon after the rediscovery of Mendel’s laws of genetic inheritance in 1900, Biffen [12] reported that yellow rust resistance in a winter wheat cultivar was controlled by a single recessive gene that segregated in a ratio of 3:1. This was the first documented case of Mendelian inheritance for disease resistance in plants. However, other workers were unable to reproduce Biffen’s results until Stakeman in 1914 [13] in Minnesota documented physiological races in the fungus with differing specificities to resistance genes in the host. These discoveries laid the foundation for breeding for disease resistance in wheat and other crops. Borlaug, who trained with Stakeman in Minnesota, will go on to work on a Rockefeller Foundation funded project in Mexico in the 1940s and usher in the Green Revolution to fight world hunger.
However, one unsolved problem remained: how do Mendel’s law of discrete inheritance factors account for continuous, quantitative or blending inheritance? Nilson-Ehle in 1909 [14] solved this riddle by an ingenious analysis of seed color inheritance in wheat where he observed ratios of 63:1, 15:1 and 3:1 red to white seeds in F2:3 families. Nilson-Ehle proposed a multifactorial hypothesis to explain red seed color inheritance; three seed color genes were segregating in some F3 families, which gave 63:1 ratio; two were segregating in others, which gave 15:1 ratio; and one gene was segregating in some that gave 3:1 ratio of red and white seeds. This led to the wide acceptance of Mendel’s laws for all types of qualitative and quantitative genetic traits and the pioneering work in wheat laid the foundation for scientific breeding for crop improvement.
4 Genome Analyzer Method, Wheat Phylogeny and Gene Pools
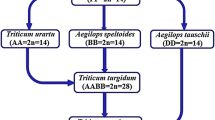
By 1915, three cultivated wheat species had been described by Schulze in Germany (cited in [3], p. 5) and Flakesberger (cited in [7]) in Russia. In an episode worthy of a suspense movie, T. Minami of Hokkaido University in Japan, in the middle of the First World War, requested these wheat seed stocks from Flakesberger in autumn 1915. Minami probably got these seeds in spring 1916 as he wrote a letter of acknowledgement in May, 1916 [7]. In 1918, a young graduate student, Tetsu Sakamura, (cited in [7]) analyzed chromosome counts of these species and discovered chromosome numbers of 2n = 14, 2n = 28 and 2n = 42 and concluded that polyploidy played a major role in wheat species phylogeny.
Sakamura also produced F1 hybrids between diploid and tetraploid species, and between tetraploid and hexaploid species. A second graduate student, H. Kihara, in 1924 (cited in [7]) analyzed chromosome pairing in triploid and pentaploid hybrids. And as often happens in science, Sax in 1922 [15] independently discovered polyploidy in wheat and also reported on the chromosome pairing in triploid and pentaploid wheat hybrids (Fig. 16.1).
First breakthrough in chromosome and genome analysis. Top panel: Homologous chromosomes pair during meiosis and this method, called the genome analyzer method, was used to elucidate chromosome and genome relationships among einkorn (T. monococcum), dicoccum (T. turgidum) and dinkel (T. aestivum) wheat species hybrids. At metaphase I (MI) of meiosis; einkorn, dicoccum and dinkel showed 7, 14 and 21 bivalents, respectively, indicating polyploidy driven speciation. The F1 hybrids between einkorn and dicoccum showed typically 3 rod and 3 ring bivalents and 9 univalents; we now know that chromosome 4A of polyploid wheat is highly rearranged and does not pair with 4A of diploid wheat. The F1 hybrids between dicoccum and dinkel wheat showed 14 bivalents and 7 univalents. The fact that chromosomes of these three species of wheat pair and recombine means that genes can be transferred from einkorn to dicoccum and dinkel, and from dicoccum to dinkel by interspecific hybridization and breeding. Figure modified with permission from [16]. Bottom panel: Current understanding of phylogeny and time line of wheat speciation [17], domestication and domestication genes (Br/br britille/nonbrittle rachis, Tg/tg tought/soft glume, q/Q speltoid/square spike)
Kihara ([3], p. 14) designated the tetraploid wheat (T. turgidum) genome as AABB and the hexaploid wheat (T. aestivum) genome as AABBDD (D as a designation of the unique genome of Dinkel wheat) and, by inference, diploid wheat (T. monococcum) as AA. Kihara reported crucial observations on the breeding behavior of pentaploid hybrids; they were semisterile and most of the progeny had chromosome numbers either close to 2n = 28, 35 or 42. This meant that, although based on F1 plant meiotic pairing of 14″ + 7′, a range of gametes (chromosome ranging from 14, 15, 16 to 21) are expected but mainly gametes with n = 14 or n = 21 functioned. This led Kihara [3] to propose the concept of the genome ([3], p. 69) as a physiological entity necessary for cell function, which was 1x = 7 unique chromosomes for wheat as mainly gametes with n = 7 or multiples of 7 such as 14 or 21 were functional.
Kihara in 1930 (cited in [7]) called phylogenetic analysis based on meiotic pairing analysis the genome analyzer method and went on to elucidate phylogenetic relationships of the wheat and Aegilops species (summarized in Kihara 1951, cited in [7]). In 1937, the drug colchicine was found to induce polyploidy by artificial chromosome doubling. McFadden and Sears in 1944 (cited in [18] produced an amphiploid by colchicine chromosome doubling of an F1 hybrid between wild emmer and Aegilops tauschii (syn Ae. squarrosa). Mcfadden and Sears [18] found that F1 hybrids between the amphiploid and bread wheat were fertile and their chromosomes paired as 21 bivalents (21″) in meiosis. Kihara in 1944 ([3], p. 82) independently produced F1 hybrids between cultivated tetraploid wheat T. persicum and Ae. tauschii and found that they were naturally fertile; he called them synthesized wheats now referred as synthetic hexaploid wheats.
The seminal and independent discoveries of Ae. tauschii as the D-genome donor of bread wheat, and artificial synthesis of bread wheat at the height of Second World War laid a scientific basis for the exploitation of tetraploid wheat and Ae. tauschii for wheat improvement. The US occupation of Japan also provided an opportunity for USDA scientist SD Salmon to procure seed of the semidwarf wheat Norin 10 (Rht1-B1, Rht2-D1), and USDA scientist Vogel at Washington State began breeding short-statured wheats (see Chap. 2).
Tetraploid wheat (T. turgidum, 2n = 28, genomes AABB) and Ae. tauschii (2n = 14, genome DD), the latter belonging to a different genus, are considered as primary gene pool species of wheat. Although there are crossability and sterility barriers because of ploidy variation, the D-genome chromosomes of Ae. tauschii and bread wheat readily pair and recombine (Riley and Chapman 1960 cited in [19]) as do the A- and B-genome chromosomes of emmer and bread wheat. McFadden (cited in [2]) made the first crosses between emmer and bread wheat in 1915, a wide-crossing method he termed “radical breeding”, and over the next 20 years bred the wheat variety ‘Hope’. Among a suite of abiotic and biotic stress traits, Hope carried a durable stem rust resistance gene Sr2.
Kihara ([3], pp. 15, 73) noted that pairing between Am-genome chromosomes of T. monococcum with the A genome of polyploid wheat was loose. Naranjo et al. 1987 (cited in [2]). discovered that chromosome 4A of polyploid wheat is highly rearranged and no longer pairs with 4A of diploid wheat. Lilienfeld and Kihara in 1934 (cited in [7], see also [3], p. 75) found that another tetraploid, T. timopheevii, had a genome composition of AAGG. Sax and Sax as early as in 1924 (cited by Linc et al. 1999 cited in [2]) reported that Ae. cylindrica had one genome in common with wheat, which was later identified as the D genome; many other polyploid species also carry D genome (Chap. 17). All these species that share partial chromosome homologies with bread wheat constitute the secondary gene pool of wheat. Doussinault et al. in 1983 (cited in [10]) transferred eyespot resistance (Pch1) from D-genome of Ae. ventricosa (DDMvMv) to chromosome 7D of wheat by homologous recombination. Later research by Barianna and McIntosh 1993, 1994 (cited in [10]) detected a cryptic transfer by spontaneous recombination involving homoeologus chromosomes 2Mv of Ae ventricosa and 2A of Ae ventricosa carrying resistance genes for rust (Lr37, Sr38, Yr17), powdery mildew, root knot nematode, wheat blast and T2A·2Mv may also boost wheat yield [20].
Kihara in 1924 (cited in [7]; see also [3], p. 14) also analyzed wheat X rye hybrids, and reported an almost complete lack of meiotic pairing between wheat and rye chromosomes; 28 univalents were observed in most cells thereby precluding genetic transfers by natural recombination. Such species constitute the tertiary gene pool of wheat. However, univalent chromosomes are prone to breakage at the centromere and spontaneous translocations involving wheat and alien chromosomes are not uncommon. Spontaneous 1B/1R substitutions and a T1BL·1RS translocation, where the long arm of chromosome 1B of wheat was translocated to the short arm of chromosome 1R of rye, was discovered in German wheat varieties by Kattermann in 1937 (cited in [16]). Wheats bred with the T1RS·1BL have a robust root system, high yield and resistance to all three rusts (Lr26, Sr31, Yr9), powdery mildew (Pm8) and some insects. This translocation was deployed with great success first, in Germany and Russia, and then worldwide from breeding efforts at CIMMYT. The Sr31 provided worldwide resistance to stem rust until Ug99 race in Uganda in 1998.
The genome analyzer method not only elucidated phylogeny of the wheat-Aegilops complex (Fig. 16.1, and Figure 1 in [2]) but also defined the wheat gene pools, thereby laying the theoretical foundation for their exploitation in wheat improvement. Borlaug used McFadden’s Hope, Vogel’s reduced height germplasm and shuttle breeding in Mexico to develop short-statured and rust-resistant varieties that launched the Green Revolution in south and west Asia beginning in the late 1960s. CIMMYT breeders bred the world’s highest yielding, second generation Green Revolution wheats based on T1B·1R. More recently, Ae, tauschii, either through direct hybridization [19] or synthetic hexaploids [21] has provided a major flux of new variation for wheat crop improvement.
5 Wheat Aneuploidy, Chromosome Mapping, and Comparative Genetics
While Kihara’s genome analysis provided a rough road map of genomic and phylogenetic relationships of wheat and Triticeae species, it revealed very little about the genetic effects of individual chromosomes. In 1936, Sears began a long-term project on wheat polyploidy by producing a large number of amphiploids from his wide-hybridization experiments. Sears (see Sears and Miller cited in [22]) selected ‘Chinese Spring’, a wheat land race from China, because of its high crossability with rye and, by inference, with other wild species. Unexpectedly, in addition to authentic wheat/rye hybrid plants, he recovered two haploid wheat plants. Upon pollination of haploids, Sears recovered 11 plants that were aneuploid 2n-1 or 2n-2 (in contrast to ploidy variation of multiples of basic genome of 1x = 7). In the progeny of one monosomic, Sears recovered a nullisomic-3B plant (missing 3B chromosome pair) that was asynaptic and isolated 17 of the possible 21 monosomic/nullisomics. Nullisomic phenotyping was used to assign a number of traits to individual chromosomes, such as the red seed trait that Ehle analyzed in 1909, awnedness, pubescence and speltoidy (q). In addition, he isolated telosomics (missing a chromosome arm), trisomics and tetrasomics and also elucidated their breeding behavior. Sears also isolated the first nullisomic-tetrasomic stock where he showed that a specific A-genome (2A) chromosome could compensate for a missing B-genome chromosome (2B) based on gametophytic and sporophytic compensation (for methodology details, see Friebe et al. 1993c cited in [2]), ushering comparative genetic analysis. Sears [23] report, “The Aneuploids of Common Wheat” on the isolation of cytogenetics stocks for the 21 chromosomes of wheat is considered the “Wheat Cytogenetics Bible” ([22]; Fig. 16.2).
Second breakthrough in chromosome and genome analysis based on aneuploid stocks. Top panel. Sears isolated many types of aneuploid stocks for targeted mapping of genes to individual chromosomes or arms bypassing genetic complexities posed by polyploidy. Three most commonly used type of aneuploid stocks and their uses are shown; such stocks are available for the 21 chromosomes of Chinese Spring wheat. Bottom panel: The aneuploid stocks in combination with deletion stocks (see Fig. 16.5) and radiation hybrid (RH) mapping [24] provide a pipeline for targeted mapping of genes as shown for trait x
In wheat breeding, one particular application was the aneuploid facilitated isolation of intercultivar wheat substitution lines that facilitated mapping of qualitative and quantitative traits to individual chromosomes (Morris and Sears 1967, cited in [22]). McFadden’s Hope cultivar genome was partitioned into 21 individual chromosome substitution lines in Chinese Spring wheat. Loegering et al. in 1957 (cited in [22]) used this material to map Hope stem rust resistance gene Sr2 on chromosome 3B. Law [25] using substitution lines, constructed a linkage map of chromosome 7B for a number of qualitative and quantitative characters. Sears cytogenetic stocks were widely shared and ensued a worldwide explosion of wheat genetics research and the first “Wheat International Genetics Symposium” (IWGS) was organized in Winnipeg in 1958 to coordinate and review wheat research at 5-year intervals. The last IWGS that was held in 2018, replaced by the International Wheat Congress to be held at 2-year intervals.
6 Chromosome Manipulation
Sears aneuploidy research also laid the foundation for directed chromosome manipulation, which he appropriately described as “chromosome engineering”, a term reserved for introgressing chromosome segments into a crop plant from different genomes of the secondary and tertiary gene pool species. These procedures are discussed in Chap. 18, see also Qi et al. [11]. O’Mara in 1940 [26] produced a set of rye chromosome additions in wheat using the first man-made crop ‘triticale’. Since then, many alien addition lines involving dozens of species have been produced (WGRC website https://www.k-state.edu/wgrc/). Wheat aneuploids and alien additions can be used to produce wheat-alien chromosome translocations as first demonstrated by Sears in 1952 (cited in [22]), and several sets have been produced [27]. Sears in 1956 (cited in [22]) also pioneered irradiation as a method to transfer alien genes into wheat and radiation hybrid mapping played a major role in the genome assembly of wheat [24].
One of the most fundamental discoveries from aneuploidy research was the identification of a pairing homoeologous gene Ph1 on 5B (Okamoto 1957, Riley and Chapman 1958, cited in [28]), which controls diploid-like pairing and disomic inheritance in polyploid wheat. Mello-Sampayo in 1971 (cited in [29]) identified a second gene, Ph2 with an intermediate effect, on 3D and encodes a mismatch repair protein MSH7-3D that inhibits homoeologous recombination. A large number of suppressors and promotors of pairing have been identified on many wheat chromosomes and in wheat species hybrids [28]. Sears in 1977 (cited in [22]) used irradiation to isolate ph1b, a deficiency mutant of Ph1. Alien chromosome transfers into wheat by induced homoeologous pairing were first demonstrated for the transfer of yellow rust resistance from Ae. comosa (Riley et al. 1968, cited in [28]) and leaf rust resistance from Agropyron (Sears 1973, cited in [22]). The ph1b-induced, homoeologous pairing, coupled with modern chromosome identification and molecular marker tools, is now the method of choice in alien gene transfer [11] (see Chap. 18).
7 Plasmon Analysis, Wheat Phylogeny and Hybrid Wheat
Kihara (1951; cited by Tsunewaki in Chapter 16 in [4]) also is credited for initiating studies on the production of nuclear-cytoplasmic substitutions and plasmon diversity in the wheat-Aegilops complex. His student, T. Tsunewaki, SS Maan in USA and Panayotov in Bulgaria, had long-running projects on alloplasmic wheat (Maan, 1975, 1991; Panytov 1983, cited in the Chapter 16 by Tsunewaki in [4]). Kihara and Tsunewaki in 1962 (cited in Chapter 16 in [4]) reported the use of alien cytoplasm for producing haploids. Tsunewaki’s group sequenced the mitochondrial and chloroplast (cp) genomes [30, 31] and demonstrated that Ae. speltoides contributed cytoplasmic genomes to both lineages of polyploid wheats (Chapter 16 in [4]). This has been validated by sequencing and haplotype analysis of cp genomes of a large number of diploid and polyploid Triticum and Aegilops species [17]. The analysis revealed that the older emmer lineage evolved 700,000 years ago compared to the timopheevii lineage that evolved 400,000 years ago (Fig. 16.1). One of the most important outcomes of plasmon analysis for wheat improvement was the discovery of a hybrid wheat production system based on Timopheevii cytoplasm (Wilson 1962, cited in [32]). Maan (cited in [32]) and his colleague Lucken at North Dakota led a major public sector effort in developing and freely sharing refined Rf gene stocks and improved A, B and R lines for a commercially viable hybrid wheat crop. Hybrid wheat received a further boost with the recent molecular cloning of fertility restoration genes Rf1-1A and Rf3-1B and sterility inducing mitochondrial orf279 transcript and molecular elucidation of their mode of action [32].
8 Protein Markers
In the mid-1960s, my fellow graduate students began using gel electrophoresis to study protein variation especially of isozymes and seed storage proteins, presumed to be direct products of genes based on the classic one gene-one protein hypothesis. Indeed, beginning with first results of aneuploid mapping of isozymes (Brewer et al. 1969 cited in [22]), especially Hart in USA and Gale and his group in the UK, identified a large set of isozyme homoeoloci that were conserved among wheat and alien homoeologous chromosomes (Chapter 12 by Hart in [4]). Thus protein markers rather than time consuming analysis of sporophytic and gametophytic compensation could be used to measure chromosome homoeologous relationships. The protein markers also found applications in wheat breeding for marker-assisted selection for linked markers for disease resistance, such as eye spot resistance (McMillen et al. 1986 cited in [10]), bread making quality (see Chap. 11) and many other traits. Protein markers gave the first indications of patterns of native wheat species diversity and wheat phylogeny, including the birth place of bread wheat (Wang et al. cited in [16]).
9 Molecular Cytogenetic Methods Provide Insights into Chromosome Substructure and Rapid Analysis of Alien Introgressions
Sears developed an exquisite cytogenetic system in wheat, yet nothing was known about the structure of individual wheat chromosomes. All chromosome identification was indirect, based on time-consuming meiotic pairing and aneuploidy analysis of F1 plants. Beginning in the late 1960s, rapid identification of somatic chromosomes in plants and animals was achieved with the discovery of Giemsa and fluorescence staining techniques (see Gill and Kimber 1974a, b cited in [1]). Simple methods were developed for DNA digestion, gel electrophoresis, cloning, labelling and mapping in Southern blots and in situ on chromosomes on a glass slide. The first experiments on wheat DNA analysis were initiated by Richard Flavell in the UK and by Rudi Appels in Australia (relevant references cited in Chapter 23 by Dvorak in [6]). We knew that the wheat genome was polyploid, but it was also large at 16 billion bp, and more than 80% was repetitive consisting of dispersed and tandemly repeated arrays (Flavell et al. 1974 and Bennett and Smith 1976 cited in Chapter 23 in [6]; Li et al. 2004 cited in [1]).
While still a graduate student at Davis, I won a grant from DF Jones Research Foundation to explore the application of new staining techniques for wheat chromosome identification for which Ernie Sears offered laboratory facilities at Missouri. Arriving in Missouri in the spring of 1973, Ernie found space for my work in Kimber’s laboratory, for Ernie did all his monumental work by himself in his large office (shared with his wife and fellow geneticist Lottie Sears), where one table was devoted to a small microscope and another with a sink for fixing wheat spikes for cytology and, incidentally, brewing coffee! I hit pay dirt soon and, based on distinctive patterns of heterochromatic bands, we cytogenetically identified the seven chromosomes of rye (Gill and Kimber 1974a cited in [1]) and the 21 chromosomes of wheat (Gill and Kimber 1974b cited in [1]). A few years later, with colleagues Friebe and Endo, we published detailed cytological maps and a nomenclature system for the 21 chromosomes of wheat (Gill et al. 1991 cited in [1]) (Fig. 16.3).
Third breakthrough in chromosome and genome analysis based on the cytogenetic identification, and resolution and description of the substructure of heterochromatic (dark staining) and euchromatic (light staining) regions of the 21 chromosomes of wheat. (Modified with permission from Gill et al. 1991, cited in [1])
In 1984, Lane Rayburn, a postdoctoral fellow in my lab from Louisiana, travelled in his cowboy attire to Stanford University (birthplace of DNA cloning) to clone “dang wheat DNA” (Rayburn and Gill 1986 cited in Chapter 23 in [6]). Rayburn isolated a clone pAs1 for identification of the D-genome chromosomes of wheat (Rayburn and Gill 1987 cited in Chapter 23 in [6]) and also developed a rapid biotin-labelling method for mapping DNA sequences on chromosomes in situ (Rayburn and Gill 1985 cited in [1]). Scharweizer and Heslop Harrison in the UK developed methods for genomic in situ hybridization (GISH), where parental genomes could be distinguished in interspecific F1 hybrids (cited in [8]). Single-copy gene sequences also can be mapped by fluorescence in situ hybridization (FISH) to discern genetic homology [34]). Thus, armed with these tools, a cytogeneticist can establish a system for any unknown species (Fig. 16.4), cytogenetically identify individual chromosomes and also discern their genomic origin and follow chromatin transfer in wide hybrids [9].
Fourth breakthrough in chromosome and genome analysis based on fluorescence in situ hybridization (FISH) mapping of DNA sequences on chromosomes. FISH and unique gene probe sets (shown as red dots) allow rapid cytogenetic identification of wheat and alien chromosomes. Wheat group 1 probe set (W1) revealed a translocation between chromosomes 1 U and 6 U of Ae umbellulata (bottom right). (Modified with permission from [34])
Advances in wide hybridization techniques (Zenkteler and Nitzse 1984, Laurie and Bennett 1986 and 1988 including the discovery of wheat/maize system for haploid breeding, cited in [8]) and new cytogenetic tools were applied to the analysis of alien introgressions [2, 5, 8, 10]. In the 1950s, wheat streak mosaic virus (WSMV), vectored by the wheat curle mite, devastated the Great Plains wheat crop. The greenhouse where I began wheat genetics research in Kansas in 1979, was built by wheat growers to tackle this menace. Daryl Wells and his group at South Dakota threw everything into the alien gene transfer tool kit, including irradiation and crosses with high-pairing Ae. speltoides, to induce alien transfer and produced a number of lines immune to WSMV from Agrotricum/wheat crosses (Lay et al. 1971 and Wells et al. 1973, 1982; cited in [10, 33]). Among this material, using C-banding and in situ hybridization, Friebe et al. 1991a (cited in [33]) identified a compensating translocation T4DL·4Ai#2S, but this line also contained another translocation T7AS-7SS·7SL (5% of 7AS of wheat and 95% Ae. speltoides 7S) that was preferentially transmitted. It took us some effort to eliminate this unwelcome alien chromosome. The T4DL·4Ai#2S harboring Wsm1, and more recent recombinants using molecular cytogenetic and DNA marker tools [11], are impacting production agriculture for control of WSMV. As usually happens, the Wsm1 recombinant also has a potent gene that provides resistance to all races of Ug99 (Yu Jin, personal communication, April 8, 2021, Manhattan, KS, USA).
In the southern Great Plains, EE Sebesta was using irradiation to transfer rye genes for greenbug (Gb6) and Hessian fly (H25) resistance to wheat. I remember visiting him in Oklahoma and he proudly showed me the irradiation gun he used to produce Amigo wheat, the donor of T1RS·1AL that does not have the adverse effect on breadmaking properties and has been widely used in production agriculture with great impact (Sebesta et al. 1995b cited in [10]). However, Sebesta was greatly devastated, for he had bred Amigo to control greenbug only to learn a new biotype had overcome the resistance. Our cytogenetic results (Lapitan et al. 1986 cited in [10]) showed that the Amigo translocation arose spontaneously by centric misdivision rather than by irradiation. Our colleague Jim Hatchett was screening another set of Sebesta’s wheat-rye irradiation materials for Hessian fly resistance. We did not work on this material while Sebesta was alive. Our posthumous analysis (Mukai et al. 1993 cited in [10]) showed that Sebesta had accomplished a rare feat and inserted a tiny bit rye chromatin harboring Hessian fly resistance H25 into a wheat chromosome and this picture made the cover of Chromosoma. I have always regretted that Sebesta was not able to appreciate the beauty of his creation during his lifetime!
One more story before I close this section. Bob McIntosh spent a mini-sabbatical in Kansas to work on mapping gene Lr45 introgressed from rye that he was unable to map by monosomic analysis. Within a few weeks, Bob determined that Lr45 was located on the translocation chromosome T2AS-2RS·2RL, consisting of a small chunk of wheat 2AS arm but half of rye 2RS arm and all of rye 2RL arm; too much alien chromatin to be useful for breeding (McIntosh 1995a, cited in [10]). Apparently, McIntosh was a victim of Murphy’s Law, for he analyzed 19 of the 21 monosomic progenies that gave noncritical ratios, except the critical monosomic 2A cross that he failed to make!
10 Chromosome Physical and DNA Marker Linkage Maps Reveal Wheat Chromosome Structural and Functional Differentiation
I spent time at UC Riverside working with Giles Waines in 1976–1977, where Lennert Johnson had amassed one of the most well-documented wild wheat collections. In Kansas, we focused our efforts on exploiting this collection for wheat improvement. Ae. tauschii proved to be a rich source of genetic diversity resistance genes, and we developed a pipeline for direct introgression using wheat/Ae. tauschii crosses and backcrosses [2, 19]. For documenting gene novelty, monosomic methods of gene mapping were cumbersome (Gill et al. 1987 cited in [6]) and we soon, in parallel with molecular cytogenetics research, began exploring RFLP (restriction fragment length polymorphism) markers for genetic mapping and tagging of useful genes.
My student Kam-Morgan was the first in our group to explore, and feel the pain and pleasure, of RFLP mapping in wheat. Because more than 90% of wheat genome consists of repetitive DNA, catching a signal of hybridization probe of a single copy clone on a X-ray film is technically demanding. But worse, 90% of the time, Lauren found that her probes did not detect polymorphism, were uninformative and wasted effort! We shifted our strategy to mapping the D-genome of wheat using an Ae. tasuchii mapping population where 75% of the probes were polymorphic. Kam-Morgan et al. in 1989 (cited in [1]) reported the first rudimentary linkage map of 5D chromosome.
Graduate student Kulvinder Gill made the first robust linkage map of Ae. tasuchii, a wild crop relative that was proving to be a gold mine for wheat improvement, using an in-house PstI-digested clone library that targets transcribed genes (Gill et al. 1991 cited in [1]). He mapped a rust resistance gene 43 cM away from marker locus D14 at the tip of chromosome arm 1DS. Postdoctoral fellow Ed Lubbers (Lubbers et al. 1991 cited in [1]) used RFLPs to analyze the structure of Ae tauschii gene pool and more recent analysis has identified two major lineages of Ae. tauschii and the birthplace of bread wheat (Wang et al. cited in [16]). RFLPs are great for comparative mapping but, for plant breeding applications, alternative breeder-friendly markers and maps were developed and of these, microsatellite marker maps, Dart arrays and more recent SNP arrays are noteworthy (see Chap. 28) (Chapter 9 by Paux and Sourdille in [6]).
I spent my sabbatical leave Down Under in Rudi Appels lab in Australia in 1986–1987 to learn the basics of DNA cloning, mapping and sequencing. As usually happens, Rudi became interested in our Ae. tauschii introgression research, and recruited Evans Lagudah to lead a GRDC project. During one of the all-important tea breaks, Sir Otto Frankel showed me a wheat chromosome banding photograph from Endo vividly demonstrating a chromosome breaking effect of an alien chromosome. Endo had visited our lab in 1981 to hone his skills in chromosome banding techniques. I immediately contacted Endo and we began a US-Japan Collaborative project on the isolation of deletion stocks (Fig. 16.5).
Fifth breakthrough in chromosome and genome analysis based on deletion stocks for targeted mapping of genes to specific regions of chromosomes. Top panel shows normal chromosome 5A (left) and 23 5A-deletion chromosomes involving the long arm from the smallest to the largest deletion (left to right). These deletion breakpoints are listed on the ideogram of 5AL on the right. The Q gene was mapped to a tiny segment of overlapping distal deletions 7 and 23, which led to the cloning of Q gene (Simons et al. 2006 cited in [1]) and many other genes in wheat. The breakpoints of 436 deletions are depicted similarly on the ideogram of 21 chromosomes of wheat. (Modified with permission from [35])
We constructed the first-generation, deletion bin-based physical maps of molecular markers for the 21 chromosomes of wheat [35]. The data provided the first glimpse of structural and functional differentiation along the chromosome length. Recombination was suppressed around the centromeric regions and gene density was low; on the contrary, recombination and gene density was high towards the chromosome ends. The deletion stocks, together with Sears’ aneuploid stocks, now could be used for targeted mapping of genes to small chromosome intervals (Fig. 16.2, bottom panel).
It was time of great molecular fervor during the 7th IWGS (1988) held in Cambridge, UK and some of us there under the leadership of Cal Qualset began discussions on the need for a coordinated international public effort for the molecular mapping of the wheat genome. The first meeting of the International Triticeae Mapping Initiative (ITMI) was held in California in 1989 and Ernie Sears attended to bless this new “wild west” of wheat research. An ITMI single-seed descent (SSD) molecular mapping population was based on a cross of Ernie’s iconic genetic model variety Chinese Spring with the first SHW genotype produced by McFadden and Sears [18]. Besides coordinating mapping efforts of the seven wheat homoeologous groups by seven research laboratories around the world, an ITMI\–NSF-funded project was launched on deletion bin mapping of the expressed portion (cDNAs) of the wheat genome using a subset of deletion stocks (Qi et al. 2003 cited in [1]). The second-generation, deletion bin-based maps of 16,000 EST loci for the 21 chromosomes of wheat (results were published in a special volume 168 of Genetics in 2004) confirmed the gene density/recombination frequency gradients and evolutionary novelty along the chromosome length (Akhunov et al. 2003a, b cited in Chapter 23 in [6]). All 64 agronomic gene tags mapped in the terminal deletion bins (Qi et. 2004 cited in [1]). My student Deven See (See et al. 2006 cited in [1]), who was a welder before he got late into science, used to say that Darwin’s workshop was located at the ends of wheat chromosomes.
The deletion bin EST maps and targeted mapping (Fig. 16.2) paved the way for cloning genes for several agronomic traits, including disease resistance genes Lr10 and Lr21, vernalization genes Vrn1 and Vrn2 and the domestication gene Q (Feuillet et al. 2003, Huang et al. 2003, Yan et al. 2003, Yan et al. 2004, Simons et al. 2006, all cited in Chapter 12 by Krattinger, Wicker and Keller in [6]). Reduced height and photoperiod genes were identified based on comparative mapping (relevant references cited in Chapters 17 and 20 in [6]). The cloned genes not only provided perfect markers for wheat breeding but also identified various alleles at each agronomic locus.
Even more important, cloned genes provide insights about their origin and evolution suggesting experimental approaches for creating new alleles, as we learned from our work with the Lr21 gene (Huang et al. 2003, Huang et al. 2009 cited in [1]). Graduate student Li Huang developed a high-resolution mapping population and, after intensive mapping, found that D14 was the closest marker. Only one plant had the D14 allele of the resistant Ae. tauschii donor but was susceptible to leaf rust. Li made a cosmid library and sequenced a 40-kb cosmid clone and it had only one disease resistance-like gene that was identical in sequence to D14. Finally, discussing the results at one of the daily WGRC ‘lunch munch’ meetings following years of frustration, we decided to forget about the exceptional plant and use the cosmid clone harboring D14 in transformation. Harold Trick gave us transgenic plants in a few months and D14 positive plants were resistant. Marker D14 was Lr21! We sequenced the exceptional F2 plant (with the resistant D14 allele but susceptible) and found that it had suffered a gene conversion and had an 800-bp DNA insertion from the susceptible parent. Sequencing of lr21 alleles, we identified an H1H1 haplotype in the spring wheat “Fielder” and an H2H2 haplotype in the winter wheat “Wichita”; intriguingly, Lr21 had a hybrid haplotype of H1H2. We crossed Fielder (H1H1) and Wichita (H2H2) and recovered the resistance allele H1H2 from intragenic recombination in a population of 5876 plants (Huang et al. 2009 cited in [1]). The recombination associated mutation rate is 170 times higher than the spontaneous mutation rate of 10−6; indeed, Darwin’s workshop is located at the ends of chromosomes!
Building on Sears’ aneuploidy based concept of comparative mapping and chromosome homoeologous relationships of wheat and alien species, Tanksley’s famous “garden blots” extended the concept of homoeology to the grass pangenome (Ahn et al. 1993 and other relevant references cited in Sorrells et al. 2003 in Chapter 17 in [6]). Thus, all grass genome information can be leveraged for the improvement of grass crops.
11 Reference Wheat Genome Sequence
As we entered the twenty-first century, Arabidopsis was sequenced in 2000 and the sequencing of rice as a model for cereal crops was underway (relevant references in Chapter 24 in [6]). In wheat, we were doing tedious chromosome walking, dirty Southerns and getting “blot” fatigue! Watching our students working with stone-age tools, I and many other wheat workers were convinced that a reference sequence and investments in wheat were needed if wheat crop technology was to stay competitive with other crops. Following an exploratory wheat genome sequencing workshop at ITMI meetings in Winnipeg in 2002, Rudi Appels and I co-organized a USDA/NSF-funded workshop in Washington DC and made a strategic plan for wheat genome sequencing [36].
The key technology component of the new strategy for mitigating disadvantages posed by a large genome size and polyploidy was the exploitation of a “chromosome genomics” platform, pioneered by Dolezel’s group in the Czech Republic (see Chapter 10 on chromosome genomics by Dolezel et al. in [6]) where they could fractionate single chromosomes and arms for sequencing or prepare DNA libraries for physical mapping. Wheat chromosomes were assigned to genome centers in 13 countries (http://www.wheatgenome.org). We had a double ditelosomic chromosome field planting in Aberdeen, ID, and sent seed material for chromosome fractionation to Dolezel’s group and from there DNA or BAC libraries went to genome centers. We were unable to get US funding for wheat genome sequencing and the leadership shifted to INRA, France under the overall leadership of Kelley Eversole (see Chapter 24 in [6]). Instead, the NSF in US chose to fund sequencing of diploid Ae. tauschii led by Jan Dvorak at UC Davis. The shot-gun sequencing papers (unanchored contig sequences, limited value) were followed by the first reference (ordered and anchored to chromosome and genetic maps, high value) sequence of chromosome 3B and survey sequences of the 21 chromosomes of wheat (IWGSC 2014 cited in [37]).
I began the chapter by recounting Sakamura’s discovery of wheat chromosome constitution and ploidy in 1918. One hundred years later, the wheat reference genome as well as the diploid D and A, the tetraploid AABB and ten elite wheat variety genome sequences have been deciphered providing information on agronomically important genomic regions (relevant references in [37, 38]). Wheat gene discovery platforms (see Chapter 13 in [6]) are driving the pace of gene discovery for precise gene tinkering using technologies such as CRISPER (see Chap. 29). Sequence-based analysis of genetic diversity, monitoring of genetic diversity during germplasm enhancement and MAS (see Chap. 28) and genomic selection (see Chap. 32) are poised to drive the efficiency and pace of genetic gain for wheat crop improvement. The applications of genomics information for conservation, management and utilization of wheat genetic resources are discussed elsewhere [16].
12 Key Concepts
The conceptual advances discussed in detail in the chapter relate to the definition of wheat gene pools defined by meiotic pairing analysis; aneuploidy facilitated genetic and comparative mapping based on gametophytic and sporophytic compensation; chromosomal structural and functional differentiation, chromosome engineering and gene novelty; wheat genome structure and function based homoeology, gene discovery and breeding; the concepts are briefly described due to space limitations and reader is highly encouraged to consult the original sources as cited through-out the chapter.
13 Conclusions
As the brief review shows, each genetic, chromosome and genomic advance facilitated the efficiency and productivity of wheat breeding. Now we are entering a new phase where one must be able to decipher the reference genomes of the parents and selected breeding lines and make selections based on masses of phenotypic and genomic data. In wide hybridization, each cross has an impact of an earthquake and one must use the concepts of homoeology to distinguish chaff from grains and cryptic transfers may be more important than the targeted transfer!
References
Raupp WJ, Friebe B (2013) Bikram Gill: cytogeneticist and wheat man. In: Plant breeding reviews 37. Wiley, pp 1–34
Gill BS, Friebe B, Raupp WJ, Wilson DL, Cox TS, Sears RG, Brown-Guedira GL, Fritz AK (2006) Wheat genetics resource center: the first 25 years. Adv. Agron. 85, Academic, pp 73–136
Kihara H (1982) Wheat studies - retrospect and prospects, vol Volume 3. Kodansha Ltd., Tokyo
Jauhar P (1996) Methods of genome analysis in plants. CRC Press
Molnár-Láng M, Ceoloni C, Doležel J (2015) Alien introgression in wheat cytogenetics, molecular biology, and genomics. Springer
Feuillet C, Muehlbauer G (2009) Genetics and genomics of the triticeae. Springer, New York
Tsunewaki K (2016) Memoir on the origin of wheat stocks used by Prof. Tetsu Sakamura, on the centennial of his discovery of the correct chromosome number and polyploidy in wheat. Genes Genet Syst 91:41–46. https://doi.org/10.1266/ggs.15-00077
Jiang J, Friebe B, Gill BS (1993) Recent advances in alien gene transfer in wheat. Euphytica 73:199–212. https://doi.org/10.1007/BF00036700
Jiang J, Gill BS (1994) Nonisotopic in situ hybridization and plant genome mapping: the first 10 years. Genome 37:717–725. https://doi.org/10.1139/g94-102
Friebe B, Jiang J, Raupp WJ, McIntosh RA, Gill BS (1996) Characterization of wheat-alien translocations conferring resistance to diseases and pests: current status. Euphytica 91:59–87. https://doi.org/10.1007/BF00035277
Qi L, Friebe B, Zhang P, Gill BS (2007) Homoeologous recombination, chromosome engineering and crop improvement. Chromosom Res 15:3–19. https://doi.org/10.1007/s10577-006-1108-8
Biffen RH (1905) Mendel’s laws of inheritance and wheat breeding. J Agric Sci 1:4–48. https://doi.org/10.1017/S0021859600000137
Stakman E (1914) A study of cereal rusts: physiological races. Minn Agric Expt Stat Bull 138
Nilson-Ehle H (1909) Einige ergebnisse von Kreungungen bei Hafer und weizen. Bot Not:257–294
Sax K (1922) Sterility in wheat hybrids. II. Chromosome behavior in partially sterile hybrids. Genetics 7:513–552
Gill B, Friebe B, Koo DH, Li W (2019) Crop species origins, the impact of domestication and the potential of wide hybridization for crop improvement. In: Zeigler R (ed) Sustaining global food security. CSIRO Publishing, Clayton, p 538
Gornicki P, Zhu H, Wang J, Challa GS, Zhang Z, Gill BS, Li W (2014) The chloroplast view of the evolution of polyploid wheat. New Phytol 204:704–714. https://doi.org/10.1111/nph.12931
McFadden ES, Sears ER (1946) The origin of Triticum spelta and its free-threshing hexaploid relatives. J Hered 37(81):107. https://doi.org/10.1093/oxfordjournals.jhered.a105590
Gill BS, Raupp WJ (1987) Direct genetic transfers from Aegilops squarrosa L. to hexaploid wheat1. Crop Sci 27. https://doi.org/10.2135/cropsci1987.0011183X002700030004x
Gao L, Koo D-H, Juliana P, Rife T, Singh D, Lemes da Silva C, Lux T, Dorn KM, Clinesmith M, Silva P, Wang X, Spannagl M, Monat C, Friebe B, Steuernagel B, Muehlbauer GJ, Walkowiak S, Pozniak C, Singh R, Stein N, Mascher M, Fritz A, Poland J (2021) The Aegilops ventricosa 2N(v)S segment in bread wheat: cytology, genomics and breeding. Theor Appl Genet 134:529–542. https://doi.org/10.1007/s00122-020-03712-y
Ogbonnaya FC, Abdalla O, Mujeeb-Kazi A, Kazi AG, Xu SS, Gosman N, Lagudah ES, Bonnett D, Sorrells ME, Tsujimoto H (2013) Synthetic hexaploids: harnessing species of the primary gene pool for wheat improvement. In: Plant breeding reviews. Wiley, pp 35–122
Riley R (1990) Ernie Sears: wheat cytogeneticist. In: 2nd International Wheat Genetics Symposium. College Ag, Univ Ext, Univ Missouri-Columbia, p 7
Sears ER (1954) The aneuploids of common wheat. Res Bull Mo Agr Exp Stn 572:59
Tiwari VK, Heesacker A, Riera-Lizarazu O, Gunn H, Wang S, Wang Y, Gu YQ, Paux E, Koo D-H, Kumar A, Luo M-C, Lazo G, Zemetra R, Akhunov E, Friebe B, Poland J, Gill BS, Kianian S, Leonard JM (2016) A whole-genome, radiation hybrid mapping resource of hexaploid wheat. Plant J 86:195–207. https://doi.org/10.1111/tpj.13153
Law CN (1967) The location of genetic factors controlling a number of quantitative characters in wheat. Genetics 56:445–461
O’mara JG (1940) Cytogenetic studies on Triticale. I. A method for determining the effects of individual Secale chromosomes on Triticum. Genetics 25:401–408
Liu W, Koo D-H, Friebe B, Gill BS (2016) A set of Triticum aestivum-Aegilops speltoides Robertsonian translocation lines. Theor Appl Genet 129:2359–2368. https://doi.org/10.1007/s00122-016-2774-3
Sears ER (1976) Genetic control of chromosome pairing in wheat. Annu Rev Genet 10:31–51. https://doi.org/10.1146/annurev.ge.10.120176.000335
Serra H, Svačina R, Baumann U, Whitford R, Sutton T, Bartoš J, Sourdille P (2021) Ph2 encodes the mismatch repair protein MSH7-3D that inhibits wheat homoeologous recombination. Nat Commun 12:803. https://doi.org/10.1038/s41467-021-21127-1
Ogihara Y, Isono K, Kojima T, Endo A, Hanaoka M, Shiina T, Terachi T, Utsugi S, Murata M, Mori N, Takumi S, Ikeo K, Gojobori T, Murai R, Murai K, Matsuoka Y, Ohnishi Y, Tajiri H, Tsunewaki K (2002) Structural features of a wheat plastome as revealed by complete sequencing of chloroplast DNA. Mol Gen Genomics 266:740–746. https://doi.org/10.1007/s00438-001-0606-9
Ogihara Y, Yamazaki Y, Murai K, Kanno A, Terachi T, Shiina T, Miyashita N, Nasuda S, Nakamura C, Mori N, Takumi S, Murata M, Futo S, Tsunewaki K (2005) Structural dynamics of cereal mitochondrial genomes as revealed by complete nucleotide sequencing of the wheat mitochondrial genome. Nucleic Acids Res 33:6235–6250. https://doi.org/10.1093/nar/gki925
Melonek J, Duarte J, Martin J, Beuf L, Murigneux A, Varenne P, Comadran J, Specel S, Levadoux S, Bernath-Levin K, Torney F, Pichon J-P, Perez P, Small I (2021) The genetic basis of cytoplasmic male sterility and fertility restoration in wheat. Nat Commun 12:1036. https://doi.org/10.1038/s41467-021-21225-0
Friebe B, Mukai Y, Dhaliwal HS, Martin TJ, Gill BS (1991) Identification of alien chromatin specifying resistance to wheat streak mosaic and greenbug in wheat germ plasm by C-banding and in situ hybridization. Theor Appl Genet 81:381–389. https://doi.org/10.1007/BF00228680
Danilova TV, Friebe B, Gill BS (2014) Development of a wheat single gene FISH map for analyzing homoeologous relationship and chromosomal rearrangements within the Triticeae. Theor Appl Genet 127:715–730. https://doi.org/10.1007/s00122-013-2253-z
Endo TR, Gill BS (1996) The deletion stocks of common wheat. J Hered 87:295–307. https://doi.org/10.1093/oxfordjournals.jhered.a023003
Gill BS, Appels R, Botha-Oberholster A-M, Buell CR, Bennetzen JL, Chalhoub B, Chumley F, Dvorák J, Iwanaga M, Keller B, Li W, McCombie WR, Ogihara Y, Quetier F, Sasaki T (2004) A workshop report on wheat genome sequencing: International Genome Research on Wheat Consortium. Genetics 168:1087–1096. https://doi.org/10.1534/genetics.104.034769
The International Wheat Genome Sequencing Consortium, Appels R, Eversole K, Stein N, Feuillet C, Keller B, Rogers J, Pozniak CJ, Choulet F, Distelfeld A, Poland J, Ronen G, Sharpe AG, Barad O, Baruch K, Keeble-Gagnère G, Mascher M, Ben-Zvi G, Josselin A-A, Himmelbach A, Balfourier F, Gutierrez-Gonzalez J, Hayden M, Koh C, Muehlbauer G, Pasam RK, Paux E, Rigault P, Tibbits J, Tiwari V, Spannagl M, Lang D, Gundlach H, Haberer G, Mayer KFX, Ormanbekova D, Prade V, Šimková H, Wicker T, Swarbreck D, Rimbert H, Felder M, Guilhot N, Kaithakottil G, Keilwagen J, Leroy P, Lux T, Twardziok S, Venturini L, Juhász A, Abrouk M, Fischer I, Uauy C, Borrill P, Ramirez-Gonzalez RH, Arnaud D, Chalabi S, Chalhoub B, Cory A, Datla R, Davey MW, Jacobs J, Robinson SJ, Steuernagel B, van Ex F, Wulff BBH, Benhamed M, Bendahmane A, Concia L, Latrasse D, Bartoš J, Bellec A, Berges H, Doležel J, Frenkel Z, Gill B, Korol A, Letellier T, Olsen O-A, Singh K, Valárik M, van der Vossen E, Vautrin S, Weining S, Fahima T, Glikson V, Raats D, Číhalíková J, Toegelová H, Vrána J, Sourdille P, Darrier B, Barabaschi D, Cattivelli L, Hernandez P, Galvez S, Budak H, Jones JDG, Witek K, Yu G, Small I, Melonek J, Zhou R, Belova T, Kanyuka K, King R, Nilsen K, Walkowiak S, Cuthbert R, Knox R, Wiebe K, Xiang D, Rohde A, Golds T, Čížková J, Akpinar BA, Biyiklioglu S, Gao L, N’Daiye A, Kubaláková M, Šafář J, Alfama F, Adam-Blondon A-F, Flores R, Guerche C, Loaec M, Quesneville H, Condie J, Ens J, Maclachlan R, Tan Y, Alberti A, Aury J-M, Barbe V, Couloux A, Cruaud C, Labadie K, Mangenot S, Wincker P, Kaur G, Luo M, Sehgal S, Chhuneja P, Gupta OP, Jindal S, Kaur P, Malik P, Sharma P, Yadav B, Singh NK, Khurana JP, Chaudhary C, Khurana P, Kumar V, Mahato A, Mathur S, Sevanthi A, Sharma N, Tomar RS, Holušová K, Plíhal O, Clark MD, Heavens D, Kettleborough G, Wright J, Balcárková B, Hu Y, Salina E, Ravin N, Skryabin K, Beletsky A, Kadnikov V, Mardanov A, Nesterov M, Rakitin A, Sergeeva E, Handa H, Kanamori H, Katagiri S, Kobayashi F, Nasuda S, Tanaka T, Wu J, Cattonaro F, Jiumeng M, Kugler K, Pfeifer M, Sandve S, Xun X, Zhan B, Batley J, Bayer PE, Edwards D, Hayashi S, Tulpová Z, Visendi P, Cui L, Du X, Feng K, Nie X, Tong W, Wang L (2018) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361:eaar7191. https://doi.org/10.1126/science.aar7191
Walkowiak S, Gao L, Monat C, Haberer G, Kassa MT, Brinton J, Ramirez-Gonzalez RH, Kolodziej MC, Delorean E, Thambugala D, Klymiuk V, Byrns B, Gundlach H, Bandi V, Siri JN, Nilsen K, Aquino C, Himmelbach A, Copetti D, Ban T, Venturini L, Bevan M, Clavijo B, Koo D-H, Ens J, Wiebe K, N’Diaye A, Fritz AK, Gutwin C, Fiebig A, Fosker C, Fu BX, Accinelli GG, Gardner KA, Fradgley N, Gutierrez-Gonzalez J, Halstead-Nussloch G, Hatakeyama M, Koh CS, Deek J, Costamagna AC, Fobert P, Heavens D, Kanamori H, Kawaura K, Kobayashi F, Krasileva K, Kuo T, McKenzie N, Murata K, Nabeka Y, Paape T, Padmarasu S, Percival-Alwyn L, Kagale S, Scholz U, Sese J, Juliana P, Singh R, Shimizu-Inatsugi R, Swarbreck D, Cockram J, Budak H, Tameshige T, Tanaka T, Tsuji H, Wright J, Wu J, Steuernagel B, Small I, Cloutier S, Keeble-Gagnère G, Muehlbauer G, Tibbets J, Nasuda S, Melonek J, Hucl PJ, Sharpe AG, Clark M, Legg E, Bharti A, Langridge P, Hall A, Uauy C, Mascher M, Krattinger SG, Handa H, Shimizu KK, Distelfeld A, Chalmers K, Keller B, Mayer KFX, Poland J, Stein N, McCartney CA, Spannagl M, Wicker T, Pozniak CJ (2020) Multiple wheat genomes reveal global variation in modern breeding. Nature 2020:1–7. https://doi.org/10.1038/s41586-020-2961-x
Acknowledgments
The author is grateful to John Raupp for excellent art work and drafting of the figures, and to John Raupp, Wanlong Li and Bernd Friebe with the editing of the manuscript and Bikram S Gill Chair for financial support.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2022 The Author(s)
About this chapter
Cite this chapter
Gill, B.S. (2022). A Century of Cytogenetic and Genome Analysis: Impact on Wheat Crop Improvement. In: Reynolds, M.P., Braun, HJ. (eds) Wheat Improvement. Springer, Cham. https://doi.org/10.1007/978-3-030-90673-3_16
Download citation
DOI: https://doi.org/10.1007/978-3-030-90673-3_16
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-90672-6
Online ISBN: 978-3-030-90673-3
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)