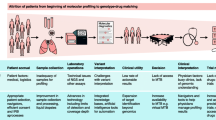
Abstract
Considerable advancements in next generation sequencing (NGS) techniques have sparked the use of comprehensive genomic profiling (CGP) as a guiding tool for precision-centered oncological treatments. The past two decades have seen the completion of the human genome project, and the consequential invention of NGS. High-throughput sequencing technologies support the discovery and commonplace use of individualized cancer treatments, specifically immune-centered checkpoint inhibitor therapies, and oncogene and tumor suppressor gene targeted therapies. Nevertheless, CGP is not commonly used in all clinical settings. This review investigates the clinically relevant applications of CGP. Studies published between the years 2000–2023 have shown substantial evidence of the benefits of integrating CGP into routine care practice, while also making important comparisons to current-standard oncological treatment strategies. Findings of a comprehensive genomic profile includes predictive, prognostic, and diagnostic biomarkers, together with somatic mutation identification which can indicate the efficacy of immunotherapies and molecularly guided therapies. This review highlights the importance of CGP in identifying driver mutations in tumors that subsequently can be effectively targeted with molecular therapeutics and lead to drug discovery, allowing for increased precision in treating tumors selectively based on their specific genetic mutations, thereby improving patient outcomes.



Similar content being viewed by others
Data availability
Data sharing is not applicable to this review article.
References
Lee H, Ross JS. The potential role of comprehensive genomic profiling to guide targeted therapy for patients with biliary cancer. Therap Adv Gastroenterol. 2017;10:507–20. https://doi.org/10.1177/1756283X17698090.
Kou T, Kanai M, Yamamoto Y, Kamada M, Nakatsui M, Sakuma T, et al. Clinical sequencing using a next-generation sequencing-based multiplex gene assay in patients with advanced solid tumors. Cancer Sci. 2017;108:1440–6. https://doi.org/10.1111/cas.13265.
Weinstein IB, Case K. The history of cancer research: introducing an AACR centennial series. Cancer Res. 2008;68:6861–2. https://doi.org/10.1158/0008-5472.CAN-08-2827.
Geoffery MC. The cell. 2nd ed. Sunderland: Sinauer Associates Inc; 2000.
Meyerson M, Gabriel S, Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet. 2010;11:685–96. https://doi.org/10.1038/nrg2841.
Calabrese C, Davidson NR, Demircioğlu D, Fonseca NA, He Y, Kahles A, et al. Genomic basis for RNA alterations in cancer. Nature. 2020;578:129–36. https://doi.org/10.1038/s41586-020-1970-0.
Comprehensive Genomic Profiling (CGP): Cancer genomic profiling benefits. Illumina n.d. https://www.illumina.com/areas-of-interest/cancer/clinical-cancer-research/cgp.html. Accessed 9 July 2021. Accessed 7 Sept 2021.
Luo W, Tian P, Wang Y, Xu H, Chen L, Tang C, et al. Characteristics of genomic alterations of lung adenocarcinoma in young never-smokers. Int J Cancer. 2018;143:1696–705. https://doi.org/10.1002/ijc.31542.
Malone ER, Oliva M, Sabatini PJB, Stockley TL, Siu LL. Molecular profiling for precision cancer therapies. Genome Med. 2020;12:1–19. https://doi.org/10.1186/s13073-019-0703-1.
Krzyszczyk P, Acevedo A, Davidoff EJ, Timmins LM, Marrero-Berrios I, Patel M, et al. The growing role of precision and personalized medicine for cancer treatment. Technology. 2018;06:79–100. https://doi.org/10.1142/S2339547818300020.
Tsimberidou AM, Fountzilas E, Nikanjam M, Kurzrock R. Review of precision cancer medicine: evolution of the treatment paradigm. Cancer Treat Rev. 2020;86: 102019. https://doi.org/10.1016/j.ctrv.2020.102019.
Pal M, Muinao T, Boruah HPD, Mahindroo N. Current advances in prognostic and diagnostic biomarkers for solid cancers: detection techniques and future challenges. Biomed Pharmacother. 2022;146: 112488. https://doi.org/10.1016/j.biopha.2021.112488.
Mateo J, Steuten L, Aftimos P, André F, Davies M, Garralda E, et al. Delivering precision oncology to patients with cancer. Nat Med. 2022;28:658–65. https://doi.org/10.1038/s41591-022-01717-2.
Rapoport BL, Troncone G, Schmitt F, Nayler SJ. Comprehensive genomic profiling. Oxford: S. Karger Publishers Ltd; 2020.
Simon R, Roychowdhury S. Implementing personalized cancer genomics in clinical trials. Nat Rev Drug Discov. 2013;12:358–69. https://doi.org/10.1038/nrd3979.
Treatment. Canadian Cancer Society 2021. www.cancer.ca. https://www.cancer.ca/en/cancer-information/diagnosis-and-treatment/treatment/?region=on. Accessed 8 Sept 2021.
Zugazagoitia J, Guedes C, Ponce S, Ferrer I, Molina-Pinelo S, Paz-Ares L. Current challenges in cancer treatment. Clin Ther. 2016;38:1551–66. https://doi.org/10.1016/j.clinthera.2016.03.026.
Looney A-M, Nawaz K, Webster RM. Tumour-agnostic therapies. Nat Rev Drug Discov. 2020;19:383–4. https://doi.org/10.1038/d41573-020-00015-1.
Haslam A, Olivier T, Tuia J, Prasad V. Umbrella review of basket trials testing a drug in tumors with actionable genetic biomarkers. BMC Cancer. 2023;23:46. https://doi.org/10.1186/s12885-022-10421-w.
Lodish H, Berk A, Zipursky S. Proto-oncogenes and tumor-suppressor genes. Molecular cell biology. 4th ed. New York: W. H. Freeman; 2000.
Jones AS. Chaos, cancer, the cellular operating system and the prediction of survival in head and neck cancer. Outcome Predict Cancer. 2007. https://doi.org/10.1016/B978-044452855-1/50007-6.
Raphael BJ, Dobson JR, Oesper L, Vandin F. Identifying driver mutations in sequenced cancer genomes: computational approaches to enable precision medicine. Genome Med. 2014;6:1–17. https://doi.org/10.1186/gm524.
Lindeman NI, Cagle PT, Aisner DL, Arcila ME, Beasley MB, Bernicker EH, et al. Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. Arch Pathol Lab Med. 2018;142:321–46. https://doi.org/10.5858/arpa.2017-0388-CP.
Toufektchan E, Toledo F. The guardian of the genome revisited: p53 downregulates genes required for telomere maintenance, DNA repair, and centromere structure. Cancers. 2018;10:135. https://doi.org/10.3390/cancers10050135.
Cadwell C, Zambetti GP. The effects of wild-type p53 tumor suppressor activity and mutant p53 gain-of-function on cell growth. Gene. 2001;277:15–30. https://doi.org/10.1016/S0378-1119(01)00696-5.
Perri F, Pisconti S, della Vittoria Scarpati G. P53 mutations and cancer: a tight linkage. Ann Transl Med 2016. https://doi.org/10.21037/atm.2016.12.40.
Zhu G, Pan C, Bei J-X, Li B, Liang C, Xu Y, et al. Mutant p53 in cancer progression and targeted therapies. Front Oncol. 2020;10: 595187. https://doi.org/10.3389/fonc.2020.595187.
Schneider K, Zelly K, Nichols KE, Garber J. Li-Fraumeni Syndrome . In: Adam M, Ardinger H, Pagon R, editors. GeneReviews, Seattle: University of Washington; 2019.
Correa H. Li-fraumeni syndrome. J Pediatr Genet. 2016. https://doi.org/10.1055/s-0036-1579759.
Liang X, Vacher S, Boulai A, Bernard V, Baulande S, Bohec M, et al. Targeted next-generation sequencing identifies clinically relevant somatic mutations in a large cohort of inflammatory breast cancer. Breast Cancer Res. 2018;20:1–12. https://doi.org/10.1186/s13058-018-1007-x.
Leading Diagnostics Companies Join Forces to Establish Access to Comprehensive Genomic Profiling Coalition. Laboratory Corporation of America Holdings n.d. https://ir.labcorp.com/news-releases/news-release-details/leading-diagnostics-companies-join-forces-establish-access. Accessed 7 Sept 2021.
Loscalzo J, Handy DE. Epigenetic modifications: basic mechanisms and role in cardiovascular disease (2013 Grover Conference Series). Pulm Circ. 2014;4:169–74. https://doi.org/10.1086/675979.
Kamińska K, Nalejska E, Kubiak M, Wojtysiak J, Żołna Ł, Kowalewski J, et al. Prognostic and predictive epigenetic biomarkers in oncology. Mol Diagn Ther. 2019;23:83–95. https://doi.org/10.1007/s40291-018-0371-7.
TruSight Oncology 500 Assay For Pan-Cancer Biomerkers in DNA and RNA . Illumina n.d. https://www.illumina.com/products/by-type/clinical-research-products/trusight-oncology-500.html. Accessed 7 Sept 2021.
Non-Small Cell Lung Cancer Targeted Drug Therapy. American Cancer Society n.d. https://www.cancer.org/cancer/lung-cancer/treating-non-small-cell/targeted-therapies.html#references. Accessed 7 Sept 2021.
Non-Small Cell Lung Cancer Treatment. NCCN Clinical Practice Guidelines in Oncology 2021. https://www.nccn.org/guidelines/guidelines-detail?category=1&id=1450. Accessed 7 Sept 2021.
Fransson Å, Glaessgen D, Alfredsson J, Wiman KG, Bajalica-Lagercrantz S, Mohell N. Strong synergy with APR-246 and DNA-damaging drugs in primary cancer cells from patients with TP53 mutant High-Grade Serous ovarian cancer. J Ovarian Res. 2016;9:1–10. https://doi.org/10.1186/s13048-016-0239-6.
Synnott NC, O’Connell D, Crown J, Duffy MJ. COTI-2 reactivates mutant p53 and inhibits growth of triple-negative breast cancer cells. Breast Cancer Res Treat. 2020;179:47–56. https://doi.org/10.1007/s10549-019-05435-1.
Pestinger V, Smith M, Sillo T, Findlay JM, Laes J-F, Martin G, et al. Use of an integrated pan-cancer oncology enrichment next-generation sequencing assay to measure tumour mutational burden and detect clinically actionable variants. Mol Diagn Ther. 2020;24:339–49. https://doi.org/10.1007/s40291-020-00462-x.
Liu L, Garbutt C, Golkaram M, Kaplan S, Martins M, Casino S, et al. Microsatellite instability testing and lynch syndrome screening for colorectal cancer patients through tumour sequencing. Ann Oncol. 2019;30: v574. https://doi.org/10.1093/annonc/mdz257.001.
Cuppens K, Froyen G, Cruys B, Geerdens E, Achten R, Vanbockrijck M, et al. P2.04-76 tumor mutational burden by TSO500 next generation sequencing panel and clinical outcome in non-small cell lung cancer. J Thorac Oncol. 2019;14:S738-9. https://doi.org/10.1016/j.jtho.2019.08.1581.
Wei B, Kang J, Kibukawa M, Arreaza G, Maguire M, Chen L, et al. Evaluation of the trusight oncology 500 assay for routine clinical testing of tumor mutational burden (TMB) and clinical utility for predicting response to pembrolizumab. J Immunother Cancer. 2020. https://doi.org/10.1136/jitc-2020-SITC2020.0080.
Koury J, Lucero M, Cato C, Chang L, Geiger J, Henry D, et al. Immunotherapies: exploiting the immune system for cancer treatment. J Immunol Res. 2018. https://doi.org/10.1155/2018/9585614.
Buchbinder EI, Desai A. CTLA-4 and PD-1 pathways. Am J Clin Oncol. 2016;39:98. https://doi.org/10.1097/COC.0000000000000239.
Alegre M-L, Frauwirth KA, Thompson CB. T-cell regulation by CD28 and CTLA-4. Nat Rev Immunol. 2001;1:220–8. https://doi.org/10.1038/35105024.
Hosseini A, Gharibi T, Marofi F, Babaloo Z, Baradaran B. CTLA-4: from mechanism to autoimmune therapy. Int Immunopharmacol. 2020;80: 106221. https://doi.org/10.1016/j.intimp.2020.106221.
Zou W, Chen L. Inhibitory B7-family molecules in the tumour microenvironment. Nat Rev Immunol. 2008;8:467–77. https://doi.org/10.1038/nri2326.
Fellner C. Ipilimumab (yervoy) prolongs survival in advanced melanoma: serious side effects and a hefty price tag may limit its use. Pharm Ther. 2012;37:503.
Seidel JA, Otsuka A, Kabashima K. Anti-PD-1 and anti-CTLA-4 therapies in cancer: mechanisms of action, efficacy, and limitations. Front Oncol. 2018;8:86. https://doi.org/10.3389/fonc.2018.00086.
Sharpe AH, Pauken KE. The diverse functions of the PD1 inhibitory pathway. Nat Rev Immunol. 2018;18:153–67. https://doi.org/10.1038/nri.2017.108.
Dong Y, Sun Q, Zhang X. PD-1 and its ligands are important immune checkpoints in cancer. Oncotarget. 2017;8:2171. https://doi.org/10.18632/oncotarget.13895.
Han Y, Liu D, Li L. PD-1/PD-L1 pathway: current researches in cancer. Am J Cancer Res. 2020;10:727.
Jiang X, Wang J, Deng X, Xiong F, Ge J, Xiang B, et al. Role of the tumor microenvironment in PD-L1/PD-1-mediated tumor immune escape. Mol Cancer. 2019;18:1–17. https://doi.org/10.1186/s12943-018-0928-4.
Lemery S, Keegan P, Pazdur R. First FDA approval agnostic of cancer site—when a biomarker defines the indication. N Engl J Med. 2017;377:1409–12. https://doi.org/10.1056/NEJMp1709968.
Smith KM, Desai J. Nivolumab for the treatment of colorectal cancer. Expert Rev Anticancer Ther. 2018;18:611–8. https://doi.org/10.1080/14737140.2018.1480942.
Ferris RL, Blumenschein G, Fayette J, Guigay J, Colevas AD, Licitra L, et al. Nivolumab for recurrent squamous-cell carcinoma of the head and neck. N Engl J Med. 2016;375:1856–67. https://doi.org/10.1056/NEJMoa1602252.
Yang Y. Cancer immunotherapy: harnessing the immune system to battle cancer. J Clin Investig. 2015;125:3335–7. https://doi.org/10.1172/JCI83871.
Yi M, Qin S, Zhao W, Yu S, Chu Q, Wu K. The role of neoantigen in immune checkpoint blockade therapy. Exp Hematol Oncol. 2018;7:1–11. https://doi.org/10.1186/s40164-018-0120-y.
Wang P, Chen Y, Wang C. Beyond tumor mutation burden: tumor neoantigen burden as a biomarker for immunotherapy and other types of therapy. Front Oncol. 2021;11: 672677. https://doi.org/10.3389/fonc.2021.672677.
Sivapiragasam A, Ashok Kumar P, Sokol ES, Albacker LA, Killian JK, Ramkissoon SH, et al. Predictive biomarkers for immune checkpoint inhibitors in metastatic breast cancer. Cancer Med. 2021;10:53–61. https://doi.org/10.1002/cam4.3550.
Guo M, Mei L, Maxwell CA. Genetic instability. Reference Module Biomed Sci. 2017. https://doi.org/10.1016/B978-0-12-801238-3.65013-4.
Cho YH, Jung SI, Hwang EC. Novel and emerging surveillance markers for bladder cancer. Bladder Cancer. 2018. https://doi.org/10.1016/B978-0-12-809939-1.00031-X.
Svrcek M, Lascols O, Cohen R, Collura A, Jonchère V, Fléjou J-F, et al. MSI/MMR-deficient tumor diagnosis: which standard for screening and for diagnosis? Diagnostic modalities for the colon and other sites: differences between tumors. Bull Cancer. 2019;106:119–28. https://doi.org/10.1016/j.bulcan.2018.12.008.
Drescher KM, Sharma P, Lynch HT. Current hypotheses on how microsatellite instability leads to enhanced survival of lynch syndrome patients. Clin Dev Immunol. 2010. https://doi.org/10.1155/2010/170432.
Yao J, Arcila ME, Ladanyi M, Hechtman JF. Pan-cancer biomarkers: changing the landscape of molecular testing. Arch Pathol Lab Med. 2021;145:692–8. https://doi.org/10.5858/arpa.2020-0513-RA.
Ballhausen A, Przybilla MJ, Jendrusch M, Haupt S, Pfaffendorf E, Seidler F, et al. The shared frameshift mutation landscape of microsatellite-unstable cancers suggests immunoediting during tumor evolution. Nat Commun. 2020;11:4740. https://doi.org/10.1038/s41467-020-18514-5.
Sahin IH, Akce M, Alese O, Shaib W, Lesinski GB, El-Rayes B, et al. Immune checkpoint inhibitors for the treatment of MSI-H/MMR-D colorectal cancer and a perspective on resistance mechanisms. Br J Cancer. 2019;121:809–18. https://doi.org/10.1038/s41416-019-0599-y.
Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–57.
André T, Shiu K-K, Kim TW, Jensen BV, Jensen LH, Punt C, et al. Pembrolizumab in microsatellite-instability—high advanced colorectal cancer. N Engl J Med. 2020;383:2207–18. https://doi.org/10.1056/NEJMoa2017699.
Andre T, Amonkar M, Norquist JM, Shiu K-K, Kim TW, Jensen BV, et al. Health-related quality of life in patients with microsatellite instability-high or mismatch repair deficient metastatic colorectal cancer treated with first-line pembrolizumab versus chemotherapy (KEYNOTE-177): an open-label, randomised, phase 3 trial. Lancet Oncol. 2021;22:665–77. https://doi.org/10.1016/S1470-2045(21)00064-4.
Ritterhouse LL. Tumor mutational burden. Cancer Cytopathol. 2019;127:735–6. https://doi.org/10.1002/cncy.22174.
Sha D, Jin Z, Budczies J, Kluck K, Stenzinger A, Sinicrope FA. Tumor mutational burden as a predictive biomarker in solid tumors. Cancer Discov. 2020;10:1808–25. https://doi.org/10.1158/2159-8290.CD-20-0522.
Goodman AM, Kato S, Bazhenova L, Patel SP, Frampton GM, Miller V, et al. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther. 2017;16:2598–608. https://doi.org/10.1158/1535-7163.MCT-17-0386.
Johnson A, Severson E, Gay L, Vergilio J, Elvin J, Suh J, et al. Comprehensive genomic profiling of 282 pediatric low- and high-grade gliomas reveals genomic drivers, tumor mutational burden, and hypermutation signatures. Oncologist. 2017;22:1478–90. https://doi.org/10.1634/theoncologist.2017-0242.
Chan TA, Yarchoan M, Jaffee E, Swanton C, Quezada SA, Stenzinger A, et al. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann Oncol. 2019;30:44–56. https://doi.org/10.1093/annonc/mdy495.
Peng M, Mo Y, Wang Y, Wu P, Zhang Y, Xiong F, et al. Neoantigen vaccine: an emerging tumor immunotherapy. Mol Cancer. 2019;18:1–14. https://doi.org/10.1186/s12943-019-1055-6.
Strickler JH, Hanks BA, Khasraw M. Tumor mutational burden as a predictor of immunotherapy response: is more always better? Clin Cancer Res. 2021;27:1236–41. https://doi.org/10.1158/1078-0432.CCR-20-3054.
Gainor JF, Rizvi H, Jimenez Aguilar E, Skoulidis F, Yeap BY, Naidoo J, et al. Clinical activity of programmed cell death 1 (PD-1) blockade in never, light, and heavy smokers with non-small-cell lung cancer and PD-L1 expression ≥50%. Ann Oncol. 2020;31:404–11. https://doi.org/10.1016/j.annonc.2019.11.015.
Chae YK, Davis AA, Raparia K, Agte S, Pan A, Mohindra N, et al. Association of tumor mutational burden with DNA repair mutations and response to anti-PD-1/PD-L1 therapy in non-small-cell lung cancer. Clin Lung Cancer. 2019;20:88–96. https://doi.org/10.1016/j.cllc.2018.09.008.
Kim JY, Kronbichler A, Eisenhut M, Hong SH, van der Vliet HJ, Kang J, et al. Tumor mutational burden and efficacy of immune checkpoint inhibitors: a systematic review and meta-analysis. Cancers. 2019;11:1798. https://doi.org/10.3390/cancers11111798.
Twomey JD, Zhang B. Cancer immunotherapy update: FDA-approved checkpoint inhibitors and companion diagnostics. AAPS J. 2021;23:1–11. https://doi.org/10.1208/s12248-021-00574-0.
Vaddepally RK, Kharel P, Pandey R, Garje R, Chandra AB. Review of indications of FDA-approved immune checkpoint inhibitors per NCCN guidelines with the level of evidence. Cancers. 2020;12:738. https://doi.org/10.3390/cancers12030738.
Milbury CA, Creeden J, Yip W-K, Smith DL, Pattani V, Maxwell K, et al. Clinical and analytical validation of FoundationOne®CDx, a comprehensive genomic profiling assay for solid tumors. PLoS ONE. 2022;17: e0264138. https://doi.org/10.1371/journal.pone.0264138.
List of Cleared or Approved Companion Diagnostic Devices . US Food and Drug Administration 2021. https://www.fda.gov/medical-devices/in-vitro-diagnostics/list-cleared-or-approved-companion-diagnostic-devices-in-vitro-and-imaging-tools. Accessed 7 Sept 2021.
Kokkat TJ, Patel MS, McGarvey D, LiVolsi VA, Baloch ZW. Archived Formalin-Fixed Paraffin-Embedded (FFPE) blocks: a valuable underexploited resource for extraction of DNA, RNA, and protein. Biopreserv Biobank. 2013;11:101–6. https://doi.org/10.1089/bio.2012.0052.
Mathieson W, Thomas GA. Why formalin-fixed, paraffin-embedded biospecimens must be used in genomic medicine: an evidence-based review and conclusion. J Histochem Cytochem. 2020;68:543–52. https://doi.org/10.1369/0022155420945050.
Nakamura Y, Taniguchi H, Ikeda M, Bando H, Kato K, Morizane C, et al. Clinical utility of circulating tumor DNA sequencing in advanced gastrointestinal cancer: SCRUM-Japan GI-SCREEN and GOZILA studies. Nat Med. 2020;26:1859–64. https://doi.org/10.1038/s41591-020-1063-5.
Luo SY, Lam DC. Oncogenic driver mutations in lung cancer. Transl Respir Med. 2013;1:1–8. https://doi.org/10.1186/2213-0802-1-6.
Birkó Z, Nagy B, Klekner Á, Virga J. Novel molecular markers in glioblastoma—benefits of liquid biopsy. Int J Mol Sci. 2020;21:7522. https://doi.org/10.3390/ijms21207522.
Delmonico L, Alves G, Bines J. Cell free DNA biology and its involvement in breast carcinogenesis. Adv Clin Chem. 2020;97:171–223. https://doi.org/10.1016/bs.acc.2019.12.006.
Ignatiadis M, Sledge GW, Jeffrey SS. Liquid biopsy enters the clinic—implementation issues and future challenges. Nat Rev Clin Oncol. 2021;18:297–312. https://doi.org/10.1038/s41571-020-00457-x.
Shohdy KS, West H. Circulating tumor DNA testing—liquid biopsy of a cancer. JAMA Oncol. 2020;6:792. https://doi.org/10.1001/jamaoncol.2020.0346.
Markou A, Tzanikou E, Lianidou E. The potential of liquid biopsy in the management of cancer patients. Semin Cancer Biol. 2022;84:69–79. https://doi.org/10.1016/j.semcancer.2022.03.013.
Stadler J-C, Belloum Y, Deitert B, Sementsov M, Heidrich I, Gebhardt C, et al. Current and future clinical applications of ctDNA in immuno-oncology. Cancer Res. 2022;82:349–58. https://doi.org/10.1158/0008-5472.CAN-21-1718.
Keller L, Belloum Y, Wikman H, Pantel K. Clinical relevance of blood-based ctDNA analysis: mutation detection and beyond. Br J Cancer. 2021;124:345–58. https://doi.org/10.1038/s41416-020-01047-5.
Chakravarty D, Solit DB. Clinical cancer genomic profiling. Nature 2021.
Thapa B, Ahmed G, Szabo A, Kamgar M, Kilari D, Mehdi M, et al. Comprehensive genomic profiling: does timing matter? Front Oncol. 2023;13:1025367. https://doi.org/10.3389/fonc.2023.1025367.
Omura T, Takahashi M, Ohno M, Miyakita Y, Yanagisawa S, Tamura Y, et al. Clinical application of comprehensive genomic profiling tests for diffuse gliomas. Cancers. 2022;14:2454. https://doi.org/10.3390/cancers14102454.
Ida H, Koyama T, Mizuno T, Sunami K, Kubo T, Sudo K, et al. Clinical utility of comprehensive genomic profiling tests for advanced or metastatic solid tumor in clinical practice. Cancer Sci. 2022;113:4300–10. https://doi.org/10.1111/cas.15586.
Acknowledgements
Maya Pankiw is supported by a Summer Studentship Award from the Department of Medicine, Mount Sinai Hospital, Toronto, Canada. The authors wish to thank Andrea Djolovic, Mount Sinai Services, for contributions to mentorship of Maya Pankiw.
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Pankiw, M., Brezden-Masley, C. & Charames, G.S. Comprehensive genomic profiling for oncological advancements by precision medicine. Med Oncol 41, 1 (2024). https://doi.org/10.1007/s12032-023-02228-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-023-02228-x