Abstract
Purpose
Triple-negative breast cancer (TNBC) currently lacks an approved targeted therapy. The tumour suppressor TP53 gene is mutated in approximately 80% of TNBC cases. COTI-2 is a third-generation thiosemicarbazone engineered for high efficacy and low toxicity which acts by reactivating mutant p53 to a WT form. The aim of this study was to investigate COTI-2 as a targeted therapy for TNBC patients.
Methods
Using a panel of 18 breast cell lines, we carried out MTT assay. p53 protein folding was determined by immunofluorescent staining with the p53 mutant-specific antibody PAb240 and the p53 WT-specific PAb1620. Surface plasmon resonance was used to determine binding affinity of COTI-2 to full length (FL) p53, and the DNA-binding domain (DBD). Flow cytometry was used to measure apoptosis.
Results
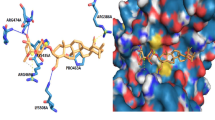
TNBC cell lines were significantly more responsive to COTI-2 than non-TNBC cell lines (p = 0.04). Furthermore, lower IC50 values were found in p53 mutant compared to p53 WT cells (p = 0.001). COTI-2 was shown to bind to FL and DBD of mutant p53. Treatment resulted in an increase in staining with PAb1620 which coincided with a decrease in staining with PAb240, suggesting refolding of the mutant protein. In addition, COTI-2 was found to induce apoptosis in TNBC cell lines.
Conclusion
We conclude that targeting mutant p53 with COTI-2 is a potential approach for treating p53-mutated TNBC.





Similar content being viewed by others
Data availability
The p53 mutational status of the cell lines used in this study was sourced from the COSMIC database, available at https://cancer.sanger.ac.uk/cell_lines.
References
Hoadley KA, Yau C, Wolf DM, Cherniack AD, Tamborero D, Ng S, Leiserson MD, Niu B, McLellan MD, Uzunangelov V, Zhang J, Kandoth C, Akbani R, Shen H, Omberg L, Chu A, Margolin AA, Van’t Veer LJ, Lopez-Bigas N, Laird PW, Raphael BJ, Ding L, Robertson AG, Byers LA, Mills GB, Weinstein JN, Van Waes C, Chen Z, Collisson EA et al (2014) Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell 158:929–1044
Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, Meyerson M, Gabriel SB, Lander ES, Getz G (2014) Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505:495–501
Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, Leiserson MD, Miller CA, Welch JS, Walter MJ, Wendl MC, Ley TJ, Wilson RK, Raphael BJ, Ding L (2013) Mutational landscape and significance across 12 major cancer types. Nature 502:333–339
Stephens PJ, Tarpey PS, Davies H, Van-Loo P, Greenman C, Wedge DC, Nik-Zainal S, Martin S, Varela I, Bignell GR, Yates LR, Papaemmanuil E, Beare D, Butler A, Cheverton A, Gamble J, Hinton J, Jia M, Jayakumar A, Jones D, Latimer C, Lau KW, McLaren S, McBride DJ, Menzies A, Mudie L, Raine K, Rad R, Chapman MS, Teague J, Easton D, Langerød A, Lee MT, Shen CY, Tee BT, Huimin BW, Broeks A, Vargas AC, Turashvili G, Martens J, Fatima A, Miron P, Chin SF, Thomas G, Boyault S, Mariani O, Lakhani SR, van de Vijver M, van’t Veer L, Foekens J, Desmedt C, Sotiriou C, Tutt A, Caldas C, Reis-Filho JS, Aparicio SA, Salomon AV, Børresen-Dale AL, Richardson AL, Campbell PJ, Futreal PA, Stratton MR, Oslo Breast Cancer Consortium (OSBREAC) (2012) The landscape of cancer genes and mutational processes in breast cancer. Nature 486:400–404
Network Cancer Genome Atlas (2012) Comprehensive molecular portraits of human breast tumours. Nature 490:61–70
Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, Turashvili G, Ding J, Tse K, Haffari G, Bashashati A, Prentice LM, Khattra J, Burleigh A, Yap D, Bernard V, McPherson A, Shumansky K, Crisan A, Giuliany R, Heravi-Moussavi A, Rosner J, Lai D, Birol I, Varhol R, Tam A, Dhalla N, Zeng T, Ma K, Chan SK, Griffith M, Moradian A, Cheng SW, Morin GB, Watson P, Gelmon K, Chia S, Chin SF, Curtis C, Rueda OM, Pharoah PD, Damaraju S, Mackey J, Hoon K, Harkins T, Tadigotla V, Sigaroudinia M, Gascard P, Tlsty T, Costello JF, Meyer IM, Eaves CJ, Wasserman WW, Jones S, Huntsman D, Hirst M, Caldas C, Marra MA, Aparicio S (2012) The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature 486:395–399
Pareja F, Geyer FC, Marchiò C, Burke KA, Weigelt B, Reis-Filho JS (2016) Triple-negative breast cancer: the importance of molecular and histologic subtyping, and recognition of low-grade variants. NPJ Breast Cancer 2:16036
Nik-Zainal S, Davies H, Staaf J, Ramakrishna M, Glodzik D, Zou X, Martincorena I, Alexandrov LB, Martin S, Wedge DC, Van Loo P, Ju YS, Smid M, Brinkman AB, Morganella S, Aure MR, Lingjærde OC, Langerød A, Ringnér M, Ahn SM, Boyault S, Brock JE, Broeks A, Butler A, Desmedt C, Dirix L, Dronov S, Fatima A, Foekens JA, Gerstung M, Hooijer GK, Jang SJ, Jones DR, Kim HY, King TA, Krishnamurthy S, Lee HJ, Lee JY, Li Y, McLaren S, Menzies A, Mustonen V, O’Meara S, Pauporté I, Pivot X, Purdie CA, Raine K, Ramakrishnan K, Rodríguez-González FG, Romieu G, Sieuwerts AM, Simpson PT, Shepherd R, Stebbings L, Stefansson OA, Teague J, Tommasi S, Treilleux I, Van den Eynden GG, Vermeulen P, Vincent-Salomon A, Yates L, Caldas C, Veer LV, Tutt A, Knappskog S, Tan BK, Jonkers J, Borg Å, Ueno NT, Sotiriou C, Viari A, Futreal PA, Campbell PJ, Span PN, Van Laere S, Lakhani SR, Eyfjord JE, Thompson AM, Birney E, Stunnenberg HG, van de Vijver MJ, Martens JW, Børresen-Dale AL, Richardson AL, Kong G, Thomas G, Stratton MR (2016) Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 534:47–54
Bykov VJN, Eriksson SE, Bianchi J, Wiman KG (2018) Targeting mutant p53 for efficient cancer therapy. Nat Rev Cancer 18:89–102
Duffy MJ, Synnott NC, Crown J (2017) Mutant p53 as a target for cancer treatment. Eur J Cancer 83:258–265
Zhao D, Tahaney WM, Mazumdar A, Savage MI, Brown PH (2017) Molecularly targeted therapies for p53-mutant cancers. Cell Mol Life Sci 74:4171–4187
Salim KY, Maleki Vareki S, Danter WR, Koropatnick J (2016) COTI-2, a novel small molecule that is active against multiple human cancer cell lines in vitro and in vivo. Oncotarget 7:41363–41379
Maleki Vareki S, Salim KY, Danter WR, Koropatnick J (2018) Novel anti-cancer drug COTI-2 synergizes with therapeutic agents and does not induce resistance or exhibit cross-resistance in human cancer cell lines. PLoS ONE 13:e0191766
Salim K, Maleki Vareki S, Danter W, Koropatnick J (2016) COTI-2, a new anticancer drug currently under clinical investigation, targets mutant p53 and negatively modulates the PI3 K/AKT/mTOR pathway. Eur J Cancer 69:S19
Lindemann A, Patel AA, Silver NL, Tang L, Liu Z, Wang L, Tanaka N, Rao X, Takahashi H, Maduka NK, Zhao M, Chen TC, Liu W, Gao M, Wang J, Frank SJ, Hittelman WN, Mills GB, Myers JN, Osman AA (2019) COTI-2, A novel thiosemicarbazone derivative, exhibits antitumor activity in HNSCC through p53-dependent and -independent mechanisms. Clin Cancer Res. https://doi.org/10.1158/1078-0432.ccr-19-0096
Ho RT, Salim KY, Lindemann A, Patel AM, Takahashi H, Wang L, Zhao M, Frank SJ, Myers J, Osman A, Lynam C, Silva AD (2018) COTI-2, a potent orally available small molecule targeting mutant p53, with promising efficacy as monotherapy and combination treatment in preclinical tumor models. J Clin Oncol 36:6040
Westin SN, Nieves-Neira W, Lynam C, Salim KY, Silva AD, Ho R, Mills GB, Coleman RL, Janku F, Matei D (2018) Safety and early efficacy signals for COTI-2, an orally available small molecule targeting p53, in a phase I trial of recurrent gynecologic cancer. In: Proceedings of the American Association for Cancer Research. http://www.abstractsonline.com/pp8/#!/4562/presentation/11184
Garrido-Castro AC, Lin NU, Polyak K (2019) Insights into molecular classifications of triple-negative breast cancer: improving patient selection for treatment. Cancer Discov 9:176–198
Synnott NC, Murray A, McGowan PM, Kiely M, Kiely PA, O’Donovan N et al (2017) Mutant p53: a novel target for the treatment of patients with triple-negative breast cancer? Int J Cancer 140(1):234–246
Synnott NC, Bauer MR, Madden S, Murray A, Klinger R, O’Donovan N et al (2018) Mutant p53 as a therapeutic target for the treatment of triple-negative breast cancer: preclinical investigation with the anti-p53 drug, PK11007. Cancer Lett 414:99–106
Joerger AC, Ang HC, Veprintsev DB, Blair CM, Fersht AR (2005) Structures of p53 cancer mutants and mechanism of rescue by second-site suppressor mutations. J Biol Chem 280:16030–16037
Chou TC (2010) Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res 70(2):440–446
Aubrey BJ, Kelly GL, Janic A, Herold MJ, Strasser A (2018) How does p53 induce apoptosis and how does this relate to p53-mediated tumour suppression? Cell Death Differ 25:104–113
Green JA, Von Euler M, Abrahmsen LB (2018) Restoration of conformation of mutant p53. Ann Oncol 29:1325–1328
Bauer MR, Joerger AC, Fersht AR (2016) 2-Sulfonylpyrimidines: mild alkylating agents with anticancer activity toward p53-compromised cells. Proc Natl Acad Sci USA 113(36):E5271–E5280
Gogna R, Madan E, Kuppusamy P, Pati U (2012) Re-oxygenation causes hypoxic tumor regression through restoration of p53 wild-type conformation and post-translational modifications. Cell Death Dis 3:e286
Milner J, Watson JV (1990) Addition of fresh medium induces cell cycle and conformation changes in p53, a tumour suppressor protein. Oncogene 5:1683–1690
Mohell N, Alfredsson J, Fransson Å, Uustalu M, Byström S, Gullbo J, Hallberg A, Bykov VJ, Björklund U, Wiman KG (2015) APR-246 overcomes resistance to cisplatin and doxorubicin in ovarian cancer cells. Cell Death Dis 6:e1794
Liu DS, Duong CP, Haupt S, Montgomery KG, House CM, Azar WJ, Pearson HB, Fisher OM, Read M, Guerra GR et al (2017) Inhibiting the system xC-/glutathione axis selectively targets cancers with mutant-p53 accumulation. Nat Commun 8:14844
Peng X, Zhang MQ, Conserva F, Hosny G, Selivanova G, Bykov VJ, Arnér ES, Wiman KG (2013) APR-246/PRIMA-1MET inhibits thioredoxin reductase 1 and converts the enzyme to a dedicated NADPH oxidase. Cell Death Dis 4:e881
Rich RL, Hoth LR, Geoghegan KF, Brown TA, LeMotte PK, Simons SP, Hensley P, Myszka DG (2002) Kinetic analysis of estrogen receptor/ligand interactions. Proc Natl Acad Sci USA 99:8562–8567
Fabian C, Tilzer L, Sternson L (1981) Comparative binding affinities of tamoxifen, 4-hydroxytamoxifen, and desmethyltamoxifen for estrogen receptors isolated from human breast carcinoma: correlation with blood levels in patients with metastatic breast cancer. Biopharm Drug Dispos 2:381–390
Lambert JM, Gorzov P, Veprintsev DB, Söderqvist M, Segerbäck D, Bergman J, Fersht AR, Hainaut P, Wiman KG, Bykov VJ (2009) PRIMA-1 reactivates mutant p53 by covalent binding to the core domain. Cancer Cell 15:376–388
Zhang Q, Bykov VJN, Wiman KG, Zawacka-Pankau J (2018) APR-246 reactivates mutant p53 by targeting cysteines 124 and 277. Cell Death Dis 9:439
Wang S, Konorev E, Kotamraju S, Joseph J, Kalivendi S, Kalyanaraman B (2004) Doxorubicin induces apoptosis in normal and tumor cells via distinctly different mechanisms. J Biol Chem 279:25535–25543
Kumar A, Palakurthi S (2013) P53 Gene therapy sensitizes resistant breast cancer cells to doxorubicin chemotherapy. Drug Deliv Lett 3:165–174
Anelli A (2003) Correlation of p53 status with outcome of neoadjuvant chemotherapy using paclitaxel and doxorubicin in stage IIIB breast cancer. Ann Oncol 14:428–432
Waks AG, Winer EP (2019) Breast cancer treatment: a review. JAMA 22(321):288–300
Acknowledgements
We thank the BREAST-PREDICT (CCRC13GAL) program of the Irish Cancer Society and the Cancer Clinical Research Trust for funding this work. We also thank Professor Alan Fersht (MRC Laboratory of Molecular Biology, Cambridge) for supplying the full length and the mutated p53-R175H DNA-binding domain of p53.
Funding
This study was funded by the Irish Cancer Society as part of the BREAST-PREDICT Collaborative Cancer Research Centre (CCRC13GAL) and the Cancer Clinical Research Trust.
Author information
Authors and Affiliations
Contributions
MJD conceived the original idea. MJD and JC supervised the project. DOC designed, carried out and analysed the surface plasmon resonance experiments. NCS carried out all other experiments and data analysis. All authors contributed to results interpretation. NCS and MJD wrote the manuscript with input from DOC.
Corresponding author
Ethics declarations
Conflict of interest
NCS, DOC and MJD have no conflict of interest to report. JC reports the following: Research Funding: Puma, GSK, Roche, BI Honoraria: Eisai, Merck Serono, Pfizer, BI, Puma, Seattle Genetics, Genomic Health, Travel grants: AbbVie, Pfizer, MSD, Stock/Ownership: Oncomark.
Ethical approval and consent to participate
No ethical approval was required for this study, due to the fact that purchased cell lines were used.
Consent to publish
All authors give consent to publish this data. No human tissue was used in this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Synnott, N.C., O’Connell, D., Crown, J. et al. COTI-2 reactivates mutant p53 and inhibits growth of triple-negative breast cancer cells. Breast Cancer Res Treat 179, 47–56 (2020). https://doi.org/10.1007/s10549-019-05435-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-019-05435-1