Abstract
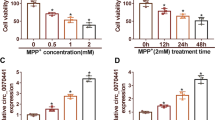
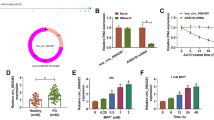
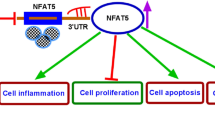
Parkinson’s disease (PD) is a common neurodegenerative disease. Circular RNAs (circRNAs) have been confirmed to regulate neurodegenerative diseases. This study was aimed to explore hsa_circ_0054220 functions in PD. MPP-stimulated SH-SY5Y cells were established as the PD cell model. PD mouse model was established by MPTP. Gene expression in cells and tissues was tested by RT-qPCR. Cell viability and apoptosis were evaluated through CCK-8 and TUNEL assays. The interactions of RNAs were determined by RNA pull-down assay, RIP assay, and luciferase reporter assay. Circ_0054220 expressed at a high level in MPP-treated SH-SY5Y cells. Circ_0054220 inhibition promoted viability and suppressed apoptosis in MPP-stimulated cells. Furthermore, we found that circ_0054220 can competitively bind to miR-145 and miR-625 to upregulate high mobility group A1 (HMGA1) expression. HMGA1 was positively regulated by circ_0054220 and overexpressed in MPP-treated cells as well as the striatum (STR), substantia nigra pars compacta (SNpc), and serum of MPTP-induced mouse model of PD. HMGA1 overexpression counteracted the function of circ_0054220 silencing on cell apoptosis. Furthermore, HMGA1 inhibition notably alleviated motor dysfunction and increased the quantity of neurons in mice resembling PD. Circ_0054220 upregulates HMGA1 by the competitive endogenous RNAs (ceRNA) pattern to promote neural impairment in PD.
Graphical Abstract









Similar content being viewed by others
Data Availability
Original data generated in this study are available from the corresponding author on reasonable request.
References
Reich, S. G., & Savitt, J. M. (2019). Parkinson’s Disease. Medical Clinics of North America, 103(2), 337–350.
Tolosa, E., et al. (2021). Challenges in the diagnosis of Parkinson’s disease. Lancet Neurology, 20(5), 385–397.
Bloem, B. R., Okun, M. S., & Klein, C. (2021). Parkinson’s Disease. Lancet, 397(10291), 2284–2303.
Erro, R., & Stamelou, M. (2017). The motor syndrome of Parkinson’s Disease. International Review of Neurobiology, 132, 25–32.
Simon, D. K., Tanner, C. M., & Brundin, P. (2020). Parkinson disease epidemiology, pathology, genetics, and pathophysiology. Clinics in Geriatric Medicine, 36(1), 1–12.
Leggio, L. (2017). microRNAs in Parkinson’s Disease: from pathogenesis to novel diagnostic and therapeutic approaches. International Journal of Molecular Sciences, 18(12), 2698
Vijiaratnam, N., et al. (2021). Progress towards therapies for disease modification in Parkinson’s disease. Lancet Neurology, 20(7), 559–572.
Kuo, M. C., et al. (2021). The role of noncoding RNAs in Parkinson’s disease: Biomarkers and associations with pathogenic pathways. Journal of Biomedical Science, 28(1), 78.
Pascale, E. (2020). Noncoding RNAs and Midbrain DA Neurons: novel molecular mechanisms and therapeutic targets in health and disease. Biomolecules, 10(9), 1269.
Liu, C. X., & Chen, L. L. (2022). Circular RNAs: Characterization, cellular roles, and applications. Cell, 185(12), 2016–2034.
Mehta, S. L., Dempsey, R. J., & Vemuganti, R. (2020). Role of circular RNAs in brain development and CNS diseases. Progress in Neurobiology, 186, 101746.
You, X., et al. (2015). Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nature Neuroscience, 18(4), 603–610.
Qian, X., et al. (2022). CircRNA_01477 influences axonal growth via regulating miR-3075/FosB/Stat3 axis. Experimental Neurology, 347, 113905.
Curry-Hyde, A., et al. (2020). Analysis of the circular transcriptome in the Synaptosomes of aged mice. Neuroscience, 449, 202–213.
Xu, K., et al. (2020). CircGRIA1 shows an age-related increase in male macaque brain and regulates synaptic plasticity and synaptogenesis. Nature Communications, 11(1), 3594.
Doxakis, E. (2022). Insights into the multifaceted role of circular RNAs: Implications for Parkinson’s disease pathogenesis and diagnosis. NPJ Parkinsons Disease, 8(1), 7.
Knowland, D., et al. (2017). Distinct ventral pallidal neural populations mediate separate symptoms of depression. Cell, 170(2), 284-297e18.
Jia, E. (2020). Transcriptomic profiling of circular RNA in different brain regions of Parkinson’s Disease in a mouse model. International Journal of Molecular Sciences, 21(8), 3006.
Kristensen, L. S., et al. (2019). The biogenesis, biology and characterization of circular RNAs. Nature Reviews Genetics, 20(11), 675–691.
Song, C., et al. (2022). Circular RNA Cwc27 contributes to Alzheimer’s disease pathogenesis by repressing Pur-α activity. Cell Death and Differentiation, 29(2), 393–406.
Dolinar, A., et al. (2019). Circular RNAs as potential blood biomarkers in amyotrophic lateral sclerosis. Molecular Neurobiology, 56(12), 8052–8062.
Hansen, T. B., et al. (2013). Natural RNA circles function as efficient microRNA sponges. Nature, 495(7441), 384–388.
Chen, L. L. (2020). The expanding regulatory mechanisms and cellular functions of circular RNAs. Nature Reviews Molecular Cell Biology, 21(8), 475–490.
Qi, X., et al. (2015). ceRNA in cancer: Possible functions and clinical implications. Journal of Medical Genetics, 52(10), 710–718.
Wang, W. (2021). circSAMD4A participates in the apoptosis and autophagy of dopaminergic neurons via the miR–29c–3p–mediated AMPK/mTOR pathway in Parkinson’s disease. Molecular Medicine Reports, 24(1), 1–10.
Liu, Q., et al. (2022). circ-Pank1 promotes dopaminergic neuron neurodegeneration through modulating miR-7a-5p/α-syn pathway in Parkinson’s disease. Cell Death and Disease, 13(5), 477.
Zhang, M., & Bian, Z. (2021). The emerging role of circular RNAs in Alzheimer’s Disease and Parkinson’s Disease. Frontiers in Aging Neuroscience, 13, 691512.
Zhang, H. (2022). The role of non-coding RNAs in the pathogenesis of Parkinson’s Disease: Recent advancement. Pharmaceuticals (Basel), 15(7), 811.
Jackson-Lewis, V., & Przedborski, S. (2007). Protocol for the MPTP mouse model of Parkinson’s disease. Nature Protocols, 2(1), 141–151.
Mustapha, M., & Mat Taib, C. N. (2021). MPTP-induced mouse model of Parkinson’s disease: A promising direction of therapeutic strategies. Bosnian Journal of Basic Medical Sciences / Udruženje Basičnih Mediciniskih Znanosti = Association of Basic Medical Sciences, 21(4), 422–433.
Li, H., et al. (2015). Anti-apoptotic effect of modified Chunsimyeolda-tang, a traditional korean herbal formula, on MPTP-induced neuronal cell death in a Parkinson’s disease mouse model. Journal of Ethnopharmacology, 176, 336–344.
Lee, S. (2021). Anti-inflammatory effects of the novel barbiturate derivative MHY2699 in an MPTP-induced mouse model of Parkinson’s Disease. Antioxidants (Basel), 10(11), 1855.
Zhou, W. Y., et al. (2020). Circular RNA: Metabolism, functions and interactions with proteins. Molecular Cancer, 19(1), 172.
Moreno-García, L. (2020). Competing endogenous RNA networks as biomarkers in neurodegenerative diseases. International Journal of Molecular Sciences, 21(24), 9582.
Akhter, R. (2018). Circular RNA and Alzheimer’s Disease. Advances in Experimental Medicine and Biology, 1087, 239–243.
Kong, F., et al. (2021). RNA-sequencing of peripheral blood circular RNAs in Parkinson disease. Medicine (Baltimore), 100(23), e25888.
Zhang, H., Wang, C., & Zhang, X. (2022). Circular RNA hsa_circ_0004381 promotes neuronal Injury in Parkinson’s Disease cell model by miR-185-5p/RAC1 axis. Neurotoxicity Research, 40(4), 1007–1019.
Cao, X., et al. (2022). Circular RNA circ_0070441 regulates MPP(+)-triggered neurotoxic effect in SH-SY5Y cells via miR-626/IRS2 axis. Metabolic Brain Disease, 37(2), 513–524.
Liu, B., Li, J., & Cairns, M. J. (2014). Identifying miRNAs, targets and functions. Briefings in Bioinformatics, 15(1), 1–19.
Mohr, A. M., & Mott, J. L. (2015). Overview of microRNA biology. Seminars in Liver Disease, 35(1), 3–11.
Bartel, D. P. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell, 116(2), 281–297.
Smillie, C. L., Sirey, T., & Ponting, C. P. (2018). Complexities of post-transcriptional regulation and the modeling of ceRNA crosstalk. Critical Reviews in Biochemistry and Molecular Biology, 53(3), 231–245.
Cheng, Q., et al. (2022). CircSV2b participates in oxidative stress regulation through mir-5107-5p-Foxk1-Akt1 axis in Parkinson’s disease. Redox Biology, 56, 102430.
Sheu-Gruttadauria, J., et al. (2019). Structural basis for target-directed MicroRNA degradation. Molecular Cell, 75(6), 1243-1255e7.
Li, X., et al. (2020). AGO2 and its partners: A silencing complex, a chromatin modulator, and new features. Critical Reviews in Biochemistry and Molecular Biology, 55(1), 33–53.
Yan, W., et al. (2016). BMP2 promotes the differentiation of neural stem cells into dopaminergic neurons in vitro via mir-145-mediated upregulation of Nurr1 expression. American Journal of Translational Research, 8(9), 3689–3699.
Xie, X., et al. (2017). miR-145-5p/Nurr1/TNF-α signaling-induced microglia activation regulates neuron injury of acute cerebral ischemic/reperfusion in rats. Frontiers in Molecular Neuroscience, 10, 383.
Ding, X. M., et al. (2019). Long non-coding RNA-p21 regulates MPP(+)-induced neuronal injury by targeting miR-625 and derepressing TRPM2 in SH-SY5Y cells. Chemico-Biological Interactions, 307, 73–81.
Reeves, R. (2000). Structure and function of the HMGI(Y) family of architectural transcription factors. Environmental Health Perspectives, 108(Suppl 5), 803–809.
Wang, L., et al. (2022). High mobility Group A1 (HMGA1): Structure, biological function, and therapeutic potential. International Journal of Biological Sciences, 18(11), 4414–4431.
Wang, Y., et al. (2019). HMGA1 in cancer: Cancer classification by location. Journal of Cellular and Molecular Medicine, 23(4), 2293–2302.
Sumter, T. F., et al. (2016). The High Mobility Group A1 (HMGA1) transcriptome in cancer and development. Current Molecular Medicine, 16(4), 353–393.
Que, T., et al. (2021). HMGA1 stimulates MYH9-dependent ubiquitination of GSK-3β via PI3K/Akt/c-Jun signaling to promote malignant progression and chemoresistance in gliomas. Cell Death and Disease, 12(12), 1147.
Manabe, T., et al. (2003). Induced HMGA1a expression causes aberrant splicing of Presenilin-2 pre-mRNA in sporadic Alzheimer’s disease. Cell Death and Differentiation, 10(6), 698–708.
Li, G., et al. (2020). HMGA1 induction of miR-103/107 forms a negative feedback loop to regulate autophagy in MPTP model of Parkinson’s Disease. Frontiers in Cellular Neuroscience, 14, 620020.
Gordon, R. (2018). Inflammasome inhibition prevents α-synuclein pathology and dopaminergic neurodegeneration in mice. Science Translational Medicine, 10(465), eaah4066.
Funding
None.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation was performed by CN. Experiments were performed by QZ, HB, and YL. Data collection and analysis was performed by CZ. The first draft of the manuscript was written by CZ and CN. All authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics Approval
This study was approved by the ethical committees of The Second Affiliated Hospital of Dalian Medical University.
Conflicting Interests
There are no conflicts of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhong, C., Zhang, Q., Bao, H. et al. Hsa_circ_0054220 Upregulates HMGA1 by the Competitive RNA Pattern to Promote Neural Impairment in MPTP Model of Parkinson’s Disease. Appl Biochem Biotechnol (2023). https://doi.org/10.1007/s12010-023-04740-2
Accepted:
Published:
DOI: https://doi.org/10.1007/s12010-023-04740-2