Abstract
Circular RNAs (circRNAs) are emerging as a novel, yet powerful player in many human diseases. They are involved in several cellular processes and are becoming a noteworthy type of biomarkers. Among other functions, circRNAs can serve as RNA sponges or as scaffolds for RNA-binding proteins. Here, we investigated a microarray expression profile of circRNAs in leukocyte samples from ALS patients and age- and sex-matched healthy controls to identify differentially expressed circRNAs. We selected 10 of them for a qPCR validation of expression on a larger set of samples, identification of their associations with clinical parameters, and evaluation of their diagnostic potential. In total, expression of 7/10 circRNAs was significant in a larger cohort of ALS patients, compared with age- and sex-matched healthy controls. Three of them (hsa_circ_0023919, hsa_circ_0063411, and hsa_circ_0088036) showed the same regulation as in microarray results. These three circRNAs also had AUC > 0.95, and sensitivity and specificity for the optimal threshold point > 90%, showing their potential for using them as diagnostic biomarkers.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease that affects both upper and lower motor neurons, resulting in muscle atrophy, speech difficulties, and respiratory insufficiency [1]. The majority of patients are classified as sporadic (SALS), while 10–15% of patients have known familial disease history (FALS) [2]. Current diagnosis of ALS is based mainly on clinical examination and it can take as much as 1 year to establish a diagnosis after the initial symptoms appeared [3]. Since patients have a mean life expectancy of 30 months [4], establishing a diagnosis represents considerable part of the disease duration. Unfortunately, approximately half of the patients receive an alternative diagnosis beforehand the ALS diagnosis [3]. Thus, reliable biomarkers are an absolute necessity for earlier and more accurate diagnosis of ALS, even more so for the diagnosis of patients with no genetic mutations or familial background. Several fluid-based biomarkers have been already proposed (for recent review on this topic, see [5]). Among them, the most promising are two proteins, neurofilament light chain (NFL) and phosphorylated neurofilament heavy chain (pNFH), that can be detected by immunoassays in cerebrospinal fluid, serum, and plasma [5]. In cases with a mutation in one of ALS-causing genes, diagnosis is confirmed by genetic testing. These mutations are associated with approximately 70% of FALS and 15% of SALS cases [2]. Mutated genes can perturb various biochemical pathways in motor neurons and lead to cell death [6]. The reason for motor neuron death in other cases remains elusive. Several epigenetic mechanisms have been already implicated in the disease development and progression [7]. Among them are also non-coding RNAs with several distinct groups of molecules—micro RNAs (miRNAs), long non-coding RNAs (lncRNAs), and small nucleolar RNAs (snRNAs) [8]. Circular RNAs (circRNAs) represent yet another class of non-coding RNAs that lately gained quite some attention [9,10,11]. Supposedly arising from back-splicing events during precursor mRNA processing, circRNAs are resistant to RNA exonucleases and thus highly stable in cells [12]. Among versatile functions of circRNAs are also miRNA sponging and RNA-binding protein (RBP) sequestration, both linked to gene regulation [13]. In the process of miRNA-sponging, each circRNA competitively binds multiple miRNAs and reduces their mRNA silencing potential [14]. Similarly, circRNAs can act as RBP-binding sites and scaffolds for protein complexes [14]. Moreover, circRNAs have been already implicated in several neurological and neurodegenerative diseases, such as glioma [15,16,17], Alzheimer’s disease [18], and Parkinson’s disease [19]. Here, circRNAs acted as miRNA sponges [16, 18, 19] or templates for protein translation [15, 17].
Considering the involvement of miRNAs in the ALS progression and potential role of circRNAs in their regulation, we wanted to determine differential expression of selected circRNAs in patients with SALS and assess their potential use as novel blood-based biomarkers for disease evaluation.
Materials and Methods
Samples
Patients were diagnosed with ALS at the Institute of Clinical Neurophysiology, University Medical Centre Ljubljana, Slovenia. Sixty patients (30 females and 30 males) were included in the study, as well as 15 age- and sex-matched healthy controls. Detailed clinical characteristics are shown in Table 1. The study was approved by the National Medical Ethics Committee of Republic of Slovenia and a written informed consent was obtained from all participants.
RNA Extraction
Peripheral blood mononuclear cells (PBMCs) were isolated from fresh blood using Ficoll density centrifugation (GE Healthcare, Sweden). Collected cells were stored in Qiazol reagent (Qiagen, Germany) at − 80 °C. Total RNA was extracted from collected cells using miRNeasy Mini Kit (Qiagen, Germany) according to the manufacturer’s instructions. The concentration and purity of total RNA were measured with NanoDrop ND-1000 (ThermoFisher, USA).
Microarray Analysis of circRNA Expression
Microarray analysis of circRNA expression was performed on a subset of 20 samples—12 patients (6 females, 6 males) and 8 age- and sex-matched controls. Total RNA from each sample was prepared for the microarray analysis according to the manufacturer’s protocol (Arraystar, USA). Briefly, total RNA was digested with RNase R (Epicentre, Inc., USA) to enrich circular RNAs. Enriched circular RNAs were amplified and transcribed into fluorescent complimentary RNA utilizing a random priming method (Arraystar Super RNA Labeling Kit; Arraystar, USA) and then hybridized onto the Arraystar Human circRNA Array V2 (8x15K, Arraystar, USA). Slides were washed and the arrays were afterwards scanned by the Agilent Scanner G2505C.
Acquired array images were analyzed using Agilent Feature Extraction software (version 11.0.1.1). Quantile normalization and subsequent data processing were performed using the R software limma package. Differentially expressed circRNAs with statistical significance between two groups were identified through volcano plot filtering. Fold change filtering was used to identify differentially expressed circRNAs between two samples. Distinguishable circRNA expression patterns among samples were identified through hierarchical clustering.
Real-time Quantitative PCR Validation of circRNA Expression
cDNA synthesis was performed on total RNA samples using SuperScript VILO Master Mix (ThermoFisher, USA). Expression levels of selected circRNAs were measured by real-time quantitative PCR (qPCR) using Sybr Select Master Mix (ThermoFisher, USA) on the Rotor Gene Q 5plex HRM platform (Qiagen, Germany) in duplicate for each sample. Primers for qPCR are shown in Table 2. Primers for RPL13A were synthesized by Qiagen (Germany) and all other were synthesized by IDT (USA). RPS17 and RPL13A were used as reference genes. The data were analyzed using the comparative cycle threshold method (2ΔΔCt).
Statistical Analysis
All experimental data were analyzed using SPSS software 24.0 (SPSS, USA). Differences in expression levels between patients and healthy controls were assessed using t test or Mann-Whitney U test, as appropriate. The correlations between circRNA expression levels and clinical data were determined by Spearman’s rank correlation. ROC curve analysis was performed to assess the diagnostic potential of statistically differentially expressed circRNAs. p value < 0.05 was considered to be statistically significant.
Results
Microarray Expression Profile
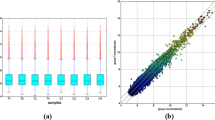
Microarray expression profile of circRNAs in ALS was performed on blood samples from 12 ALS patients and 8 age- and sex-matched healthy controls using Arraystar Human circRNA Array Analysis. Hierarchical clustering and subsequent heatmap visualization of circRNA expression levels in samples showed distinguishable expression patterns among healthy controls and ALS patients (Fig. 1a). Moreover, we analyzed differences in expression levels using volcano plot (Fig. 1b) and identified 425 differentially expressed circRNAs when comparing ALS patients and healthy controls (circRNAs with fold change > 1.5 and p value < 0.05). Of them, 274 were upregulated and 151 were downregulated in ALS patients.
Summary of microarray expression profile. a Hierarchical clustering and heatmap visualization of circRNA expression levels. P, ALS patient; C, healthy control. b Volcano plot representation of differentially expressed circRNAs (red points; p value < 0.05 and fold change > 1.5). Two hundred seventy-four of them were upregulated and 151 were downregulated when comparing ALS patients and healthy controls
Following the initial microarray analysis, we selected 10 circRNAs for qPCR validation of their expression. Selection criteria included, but were not limited to, p value (< 0.04), fold change (> 1.8), genomic location (exonic), and function of the hosting gene. We evaluated the function of the hosting gene and its potential involvement in ALS through literature and database search (ALSoD [20]: http://alsod.iop.kcl.ac.uk/; Ensembl [21]: release 94). Summary of these criteria for selected circRNAs is shown in Table 3.
qPCR Validation of circRNA Expression
Microarray expression results were validated with qPCR on blood samples from 60 ALS patients and 15 age- and sex-matched healthy controls. Of 10 selected circRNAs, we got 6 significantly upregulated circRNAs (hsa_circ_0000567, hsa_circ_0005218, hsa_circ_0035796, hsa_circ_0043138, hsa_circ_0063411, and hsa_circ_0088036) and 1 significantly downregulated circRNA (hsa_circ_0023919) in ALS patients. hsa_circ_0005896 and hsa_circ_0001173 showed no significant difference in expression between ALS patients and healthy controls and expression levels of hsa_circ_0073647 were not detectable in either ALS patients or healthy controls (Fig. 2). Among significantly dysregulated circRNAs, hsa_circ_0063411 and hsa_circ_0088036 were upregulated in both microarray and qPCR analyses, while hsa_circ_0023919 was downregulated in both experiments. Four circRNAs (hsa_circ_0000567, hsa_circ_0005218, hsa_circ_0035796, and hsa_circ_0043138) showed discrepancy between the results from the two experiments as they were downregulated in microarray analysis and upregulated in qPCR results.
Expression of selected circRNAs in ALS patients and healthy controls. Relative expression levels of each circRNA in ALS patients (n = 60) and healthy controls (n = 15) are represented with box plot. Upregulated circRNAs are shown in green, downregulated circRNAs in red, and circRNAs with non-significant differences in expression in blue; significant difference in expression levels is denoted as * (p < 0.05) or *** (p < 0.001); n. s., non-significant
Associations Between Clinical Variables and circRNA Expression
The Spearman rank correlation test was performed to assess potential associations between circRNA expression and clinical variables. As shown in Table 4, the expression levels of hsa_circ_0000567 and hsa_circ_0088036 were negatively associated with age, both at the time of blood collection and at the time of disease onset. There was no association between the expression of either of these two circRNAs and the age at the time of blood collection in healthy controls (hsa_circ_0000567: Spearman’s rho = − 0.020, p = 0.944; hsa_circ_0088036: Spearman’s rho = 0.263, p = 0.725). Another negative association was found between the expression of hsa_circ_0023919 and the age, however, only at the time of blood collection. Similarly, there was no association between hsa_circ_0023919 expression and the age at the time of blood collection in healthy controls (Spearman’s rho = − 0.059, p = 0.834). The expression levels of hsa_circ_0063411 and hsa_circ_0005218 were positively associated with the level of functional impairment in patients and the expression of hsa_circ_0063411 was negatively associated also with the disease duration and survival time. Also, the expression levels of several circRNA were positively correlated with each other.
Determination of circRNA Diagnostic Potential by ROC Curve Analysis
We performed receiver operating characteristics (ROC) curve analysis to evaluate the diagnostic potential of selected circRNAs (Fig. 3). Of 7 circRNAs with statistical difference in expression levels between cases and controls, three (hsa_circ_0023919, red; hsa_circ_0088036, green; hsa_circ_0063411, blue) had area under the curve (AUC) over 0.950 (Fig. 3a, b). These circRNAs also had outstanding specificity and sensitivity at the optimal threshold point—over 90% (Fig. 3c). AUC values for other circRNAs varied between 0.623 and 0.894 (dashed gray lines) (Fig. 3a, c).
ROC curves for the circRNAs with statistically significant difference in expression. Full colored lines represent circRNAs with AUC > 0.950, dashed gray lines represent other circRNAs. a ROC curves for upregulated circRNAs. b ROC curve for downregulated circRNA. c Details of shown ROC curves. AUC, area under the curve, SE, standard error for AUC, 95% C. I., 95% confidence interval for AUC. Optimal threshold point was determined as the point on the curve with minimal distance to the ideal point (sensitivity = 1 and specificity = 1)
Discussion
circRNAs are widely expressed in several human tissues [32, 33] and have been already implicated in numerous developmental and physiological processes—myogenesis [34], synaptogenesis [35], and cell growth [36]. Inevitably, they are implicated also in pathological processes—tumorigenesis [16, 37], abnormal mRNA splicing [38], and neurodegeneration [18, 19]. However, no ALS-associated circRNA has been identified yet in human samples. Here, we present the first circRNA differential expression analysis in leukocyte samples from patients with amyotrophic lateral sclerosis. Muscle and nervous tissues are the most affected tissues in ALS; however, blood samples are easier to obtain and thus more suitable for diagnostics if reliable biomarkers exist.
Microarray-based circRNA expression profiling on a representative subset of samples revealed 274 upregulated and 151 downregulated circRNAs between ALS patients and healthy controls. Based on the microarray results and our estimated relevant function of a hosting gene in ALS, we selected 10 circRNAs for further validation of expression on a larger set of samples. In total, expression of 7 of 10 selected circRNAs was significant between ALS samples and healthy controls. Four of them (hsa_circ_0000567, hsa_circ_0023919, hsa_circ_0063411, and hsa_circ_0088036) showed the highest significance as well as clinical relevance. In addition, 3 of them (hsa_circ_0023919, hsa_circ_0063411, and hsa_circ_0088036) also showed identical regulation in both microarray and qPCR results.
hsa_circ_0000567 is located in SETD3 gene, product of which is histone methyltransferase that regulates muscle differentiation in mouse [22]. This circRNA was downregulated in microarray analysis; however, qPCR results showed significant upregulation in ALS cases. According to Morey et al. [39], correlation of microarray and qPCR results is gene specific and can vary considerably. Particularly in the cases where microarray showed downregulation and qPCR validation failed to confirm that, the result discrepancy may be due to variability in array spot intensity or due to increased sample size in qPCR analysis.
hsa_circ_0023919 is located in PICALM gene that is involved in clathrin-mediated endocytosis at neuromuscular junctions [26] and single nucleotide polymorphism upstream of the gene has been associated with Alzheimer’s disease [40]. This circRNA was downregulated in microarray analysis and qPCR results also confirmed this. hsa_circ_0023919 sequence contains two binding sites for hsa-miR-9 (imperfect binding site between 61 and 67 bp and 7mer-m8 binding site between 142 and 148 bp) [41]. Upregulation of miR-9 was confirmed in both mouse model of ALS [42] and in human blood samples of ALS patients [43]. By all means, further functional studies are necessary to investigate the potential association between hsa_circ_0023919 and miR-9 in ALS.
hsa_circ_0063411’s host gene, TNRC6B, guides Ago-mediated gene silencing [29]. This circRNA was upregulated in both microarray and qPCR analyses. There is no evidence yet on the role of this circRNA in any biological or pathological process. However, it contains one binding site for hsa-miR-647 (7mer-m8 site between 680 and 686 bp) [41]. Some connection between hsa-miR-647 and ALS was already detected in spinal cord samples from ALS patients where miRNA-647 was found downregulated [44]. In order to elucidate the potential associations between circ_0063411 and hsa-miR-647 expression in ALS patients and their common roles in ALS disease initiation and progression, further studies are necessary.
hsa_circ_0088036 is located in SUSD1 gene that is potentially associated with ALS [31]. hsa_circ_0088036 was, like hsa_circ_0063411, upregulated in microarray and qPCR experiments. Previous study showed that hsa_circ_0088036 (also known as hsa_circRNA_104871) was significantly upregulated in PBMCs from patients with rheumatoid arthritis and may serve as a potential biomarker for its diagnosis [45]. Since this circRNA was significantly upregulated also in this study and diseases have no common cause, we could speculate that hsa_circ_0063411 might somehow be involved in the immune response. Nevertheless, further studies are necessary to confirm that.
All of selected circRNAs (Table 3) have one or more predicted binding sites for several RBPs. Two of them, AGO2 and EIF4A3, can bind to all of the selected circRNAs with the exception of hsa_circ_0035796 that has binding sites only for EIF4A3. Despite potential role of circRNA to serve as scaffolds for protein complexes [14], the variability in the number of binding sites and presence in circRNAs of various origins and functions indicate other potential explanations. As Chen et al. [46] showed, RBPs are involved in discriminating between endogenous and exogenous circRNAs and abolishing immune response to endogenous circRNAs. Origin discrimination is based on intronic sequences that are involved in splicing and circularization and associated with splicing complexes, part of which is also EIF4A3 [46].
Another RNA-binding protein, fused in sarcoma (FUS), has been recognized as an important modulator of circRNA expression [47]. Errichelli et al. observed an overall downregulation of circRNA expression in FUS−/− mice and expression was dysregulated also in FUSR521C and FUSP525L human-induced pluripotent stem cell-derived motor neurons. Since cognate linear transcripts showed no significant alteration in expression levels, circRNA deregulation can be attributed to altered splicing dynamics due to mutated or absent FUS. Whether this is the reason for altered circRNA expression also in human tissues, it remains to be determined. These findings could have considerable implications for further research on circRNAs in ALS as mutations in FUS have been found in 5% of ALS cases [2].
With the Spearman rank correlation test, we evaluated the potential associations between circRNA expression and clinical data or correlations between expression levels of each circRNA. We found that three circRNAs (hsa_circ_0000567, hsa_circ_0023919, and hsa_circ_0088036) were negatively associated with the age of ALS patient at the time of blood collection and two of them (hsa_circ_0000567 and hsa_circ_0088036) were also negatively associated with the age at the disease onset. None of these circRNAs were associated with the age at the time of blood collection in healthy controls, suggesting that this association is disease specific. hsa_circ_0005218 was positively correlated with the level of functional impairment in ALS patients and hsa_circ_0063411 was negatively correlated with disease duration and survival time. Verification of clinically relevant associations is needed to exclude potential influence of other clinical conditions. Moreover, expression of several circRNAs was positively correlated with each other, indicating potential involvement of these circRNAs in similar biological processes and/or co-regulation. Extensive functional studies are of course needed to evaluate these indications.
Through ROC curve analysis, we identified hsa_circ_0023919, hsa_circ_0088036, and hsa_circ_0063411 as potential blood-based biomarkers for ALS. All of them had AUC values above 0.95. Moreover, at the optimal threshold point, each of them had both sensitivity and specificity above 90%. Among already discovered fluid-based biomarkers for ALS are the most promising two protein biomarkers, NFL and pNFH. However, they do not reach such sensitivity and specificity in serum or plasma samples [5] as have reached circRNAs in this study. Therefore, we could speculate that circRNAs hsa_circ_0023919, hsa_circ_0088036, and hsa_circ_0063411 could possess great clinical relevance in ALS. However, studies with increased sample size and more diverse set of controls are needed to justify this. Furthermore, comparison with other neurodegenerative diseases is necessary in order to investigate ALS disease specificity.
In conclusion, to our knowledge, this is the first study of circRNA expression profile in human samples of ALS. It provides a broad framework for further functional studies on the role of circRNAs in ALS. This might help to improve our understanding about the molecular mechanisms in ALS. This work also revealed promising diagnostic potential of circRNAs. We think therefore that circRNAs and their association with ALS are definitely worth to be further investigated.
Abbreviations
- ALS:
-
Amyotrophic lateral sclerosis
- ALS-FRS-R:
-
ALS functional rating scale revised
- AUC:
-
Area under the curve
- circRNA:
-
Circular RNA
- FALS:
-
Familial ALS
- NFL:
-
Neurofilament light chain
- PBMCs:
-
Peripheral blood mononuclear cells
- pNFH:
-
Phosphorylated neurofilament heavy chain
- RBP:
-
RNA-binding protein
- ROC:
-
Receiver operating characteristics
- SALS:
-
Sporadic ALS
- snRNAs:
-
Small nucleolar RNAs
References
Hobson EV, Harwood CA, McDermott CJ, Shaw PJ (2016) Clinical aspects of motor neurone disease. Medicine 44(9):552–556. https://doi.org/10.1016/j.mpmed.2016.06.004
Chia R, Chiò A, Traynor BJ (2018) Novel genes associated with amyotrophic lateral sclerosis: diagnostic and clinical implications. Lancet Neurol 17(1):94–102. https://doi.org/10.1016/S1474-4422(17)30401-5
Paganoni S, Macklin EA, Lee A, Murphy A, Chang J, Zipf A, Cudkowicz M, Atassi N (2014) Diagnostic timelines and delays in diagnosing amyotrophic lateral sclerosis (ALS). Amyotroph Lateral Scler Frontotemporal Degeneration 15(5–6):453–456. https://doi.org/10.3109/21678421.2014.903974
Hardiman O, Al-Chalabi A, Brayne C, Beghi E, van den Berg LH, Chio A, Martin S, Logroscino G et al (2017) The changing picture of amyotrophic lateral sclerosis: lessons from European registers. J Neurol Neurosurg Psychiatry 88:557–563. https://doi.org/10.1136/jnnp-2016-314495
Vu LT, Bowser R (2017) Fluid-based biomarkers for amyotrophic lateral sclerosis. Neurotherapeutics 14(1):119–134. https://doi.org/10.1007/s13311-016-0503-x
Weishaupt JH, Hyman T, Dikic I (2016) Common molecular pathways in amyotrophic lateral sclerosis and frontotemporal dementia. Trends Mol Med 22(9):769–783. https://doi.org/10.1016/j.molmed.2016.07.005
Dolinar A, Ravnik-Glavač M, Glavač D (2018) Epigenetic mechanisms in amyotrophic lateral sclerosis: a short review. Mech Ageing Dev 174:103–110. https://doi.org/10.1016/j.mad.2018.03.005
Salta E, De Strooper B (2017) Noncoding RNAs in neurodegeneration. Nat Rev Neurosci 18:627–640. https://doi.org/10.1038/nrn.2017.90
Chen LL (2016) The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol 17(4):205–211. https://doi.org/10.1038/nrm.2015.32
Rong D, Sun H, Li Z, Liu S, Dong C, Fu K, Tang W, Cao H (2017) An emerging function of circRNA-miRNAs-mRNA axis in human diseases. Oncotarget 8(42):73271. https://doi.org/10.18632/oncotarget.19154
Xie L, Mao M, Xiong K, Jiang B (2017) Circular RNAs: a novel player in development and disease of the central nervous system. Front Cell Neurosci 11:354. https://doi.org/10.3389/fncel.2017.00354
Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE (2013) Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19(2):141–157. https://doi.org/10.1261/rna.035667.112
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD et al (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495:333–338. https://doi.org/10.1038/nature11928
van Rossum D, Verheijen BM, Pasterkamp RJ (2016) Circular RNAs: novel regulators of neuronal development. Front Mol Neurosci 9:74. https://doi.org/10.3389/fnmol.2016.00074
Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F, Huang N, Yang X et al (2018) Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. JNCI: J Natl Cancer Inst 110(3):304–315. https://doi.org/10.1093/jnci/djx166
Xu H, Zhang Y, Qi L, Ding L, Jiang H, Yu H (2018) NFIX circular RNA promotes glioma progression by regulating miR-34a-5p via Notch signaling pathway. Front Mol Neurosci 11:225. https://doi.org/10.3389/fnmol.2018.00225
Zhang M, Huang N, Yang X, Luo J, Yan S, Xiao F, Chen W, Gao X et al (2018) A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene 37(13):1805–1814. https://doi.org/10.1038/s41388-017-0019-9
Zhao Y, Alexandrov P, Jaber V, Lukiw W (2016) Deficiency in the ubiquitin conjugating enzyme UBE2A in Alzheimer’s disease (AD) is linked to deficits in a natural circular miRNA-7 sponge (circRNA; ciRS-7). Genes 7(12):116. https://doi.org/10.3390/genes7120116
Sang Q, Liu X, Wang L, Qi L, Sun W, Wang W, Sun Y, Zhang H (2018) CircSNCA downregulation by pramipexole treatment mediates cell apoptosis and autophagy in Parkinson’s disease by targeting miR-7. Aging 10(6):1281–1293. https://doi.org/10.18632/aging.101466
Abel O, Powell JF, Andersen PM, Al-Chalabi A (2012) ALSoD: a user-friendly online bioinformatics tool for amyotrophic lateral sclerosis genetics. Hum Mutat 33(9):1345–1351. https://doi.org/10.1002/humu.22157
Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, Billis K, Cummins C et al (2018) Ensembl 2018. Nucleic Acids Res 46(D1):D754–D761. https://doi.org/10.1093/nar/gkx1098
Eom GH, Kim KB, Kim JH, Kim JY, Kim JR, Kee HJ, Kim DW, Choe N et al (2011) Histone methyltransferase SETD3 regulates muscle differentiation. J Biol Chem 286(40):34733–34742. https://doi.org/10.1074/jbc.M110.203307
Nishimura AL, Mitne-Neto M, Silva HC, Richieri-Costa A, Middleton S, Cascio D, Kok F, Oliveira JR et al (2004) A mutation in the vesicle-trafficking protein VAPB causes late-onset spinal muscular atrophy and amyotrophic lateral sclerosis. Am J Hum Genet 75(5):822–831. https://doi.org/10.1086/425287
Huttlin EL, Bruckner RJ, Paulo JA, Cannon JR, Ting L, Baltier K, Colby G, Gebreab F et al (2017) Architecture of the human interactome defines protein communities and disease networks. Nature 545:505–509. https://doi.org/10.1038/nature22366
Lotti F, Imlach Wendy L, Saieva L, Beck Erin S, Hao Le T, Li Darrick K, Jiao W, Mentis George Z et al (2012) An SMN-dependent U12 splicing event essential for motor circuit function. Cell 151(2):440–454. https://doi.org/10.1016/j.cell.2012.09.012
Tebar F, Bohlander SK, Sorkin A (1999) Clathrin assembly lymphoid myeloid leukemia (CALM) protein: localization in endocytic-coated pits, interactions with clathrin, and the impact of overexpression on clathrin-mediated traffic. Mol Biol Cell 10(8):2687–2702. https://doi.org/10.1091/mbc.10.8.2687
Bachiller S, Rybkina T, Porras-Garcia E, Perez-Villegas E, Tabares L, Armengol JA, Carrion AM, Ruiz R (2015) The HERC1 E3 ubiquitin ligase is essential for normal development and for neurotransmission at the mouse neuromuscular junction. Cell Mol Life Sci 72(15):2961–2971. https://doi.org/10.1007/s00018-015-1878-2
Ticozzi N, Vance C, Leclerc AL, Keagle P, Glass JD, McKenna-Yasek D, Sapp PC, Silani V et al (2011) Mutational analysis reveals the FUS homolog TAF15 as a candidate gene for familial amyotrophic lateral sclerosis. Am J Med Genet B Neuropsychiatr Genet 156B(3):285–290. https://doi.org/10.1002/ajmg.b.31158
Nishi K, Nishi A, Nagasawa T, Ui-Tei K (2013) Human TNRC6A is an Argonaute-navigator protein for microRNA-mediated gene silencing in the nucleus. RNA (New York, NY) 19(1):17–35. https://doi.org/10.1261/rna.034769.112
Perez-Branguli F, Zagar Y, Shanley DK, Graef IA, Chédotal A, Mitchell KJ (2016) Reverse signaling by Semaphorin-6A regulates cellular aggregation and neuronal morphology. PLoS One 11(7):e0158686. https://doi.org/10.1371/journal.pone.0158686
Schymick JC, Scholz SW, Fung HC, Britton A, Arepalli S, Gibbs JR, Lombardo F, Matarin M et al (2007) Genome-wide genotyping in amyotrophic lateral sclerosis and neurologically normal controls: first stage analysis and public release of data. Lancet Neurol 6(4):322–328. https://doi.org/10.1016/s1474-4422(07)70037-6
Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S, Hanan M, Behm M et al (2015) Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell 58(5):870–885. https://doi.org/10.1016/j.molcel.2015.03.027
Maass PG, Glažar P, Memczak S, Dittmar G, Hollfinger I, Schreyer L, Sauer AV, Toka O et al (2017) A map of human circular RNAs in clinically relevant tissues. J Mol Med 95(11):1179–1189. https://doi.org/10.1007/s00109-017-1582-9
Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, Fatica A, Santini T et al (2017) Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell 66(1):22–37.e29. https://doi.org/10.1016/j.molcel.2017.02.017
Chen BJ, Huang S, Janitz M (2018) Changes in circular RNA expression patterns during human foetal brain development. Genomics. https://doi.org/10.1016/j.ygeno.2018.04.015
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D et al (2016) Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun 7:11215. https://doi.org/10.1038/ncomms11215
Shang X, Li G, Liu H, Li T, Liu J, Zhao Q, Wang C (2016) Comprehensive circular RNA profiling reveals that hsa_circ_0005075, a new circular RNA biomarker, is involved in hepatocellular carcinoma development. Medicine (Baltimore) 95(22):e3811. https://doi.org/10.1097/md.0000000000003811
Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S et al (2014) circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 56(1):55–66. https://doi.org/10.1016/j.molcel.2014.08.019
Morey JS, Ryan JC, Van Dolah FM (2006) Microarray validation: factors influencing correlation between oligonucleotide microarrays and real-time PCR. Biol Proced Online 8:175–193. https://doi.org/10.1251/bpo126
Santos-Reboucas CB, Goncalves AP, Dos Santos JM, Abdala BB, Motta LB, Laks J, de Borges MB, de Rosso ALZ et al (2017) rs3851179 polymorphism at 5′ to the PICALM gene is associated with Alzheimer and Parkinson diseases in Brazilian population. NeuroMolecular Med 19(2–3):293–299. https://doi.org/10.1007/s12017-017-8444-z
Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M (2016) CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol 13(1):34–42. https://doi.org/10.1080/15476286.2015.1128065
Zhou F, Guan Y, Chen Y, Zhang C, Yu L, Gao H, Du H, Liu B et al (2013) miRNA-9 expression is upregulated in the spinal cord of G93A-SOD1 transgenic mice. Int J Clin Exp Pathol 6(9):1826–1838
Vrabec K, Boštjančič E, Koritnik B, Leonardis L, Dolenc Grošelj L, Zidar J, Rogelj B, Glavač D et al (2018) Differential expression of several miRNAs and the host genes AATK and DNM2 in leukocytes of sporadic ALS patients. Front Mol Neurosci 11:106. https://doi.org/10.3389/fnmol.2018.00106
Campos-Melo D, Droppelmann CA, He Z, Volkening K, Strong MJ (2013) Altered microRNA expression profile in amyotrophic lateral sclerosis: a role in the regulation of NFL mRNA levels. Mol Brain 6(1):26. https://doi.org/10.1186/1756-6606-6-26
Ouyang Q, Wu J, Jiang Z, Zhao J, Wang R, Lou A, Zhu D, Shi GP et al (2017) Microarray expression profile of circular RNAs in peripheral blood mononuclear cells from rheumatoid arthritis patients. Cell Physiol Biochem 42(2):651–659. https://doi.org/10.1159/000477883
Chen YG, Kim MV, Chen X, Batista PJ, Aoyama S, Wilusz JE, Iwasaki A, Chang HY (2017) Sensing self and foreign circular RNAs by intron identity. Mol Cell 67(2):228–238.e225. https://doi.org/10.1016/j.molcel.2017.05.022
Errichelli L, Dini Modigliani S, Laneve P, Colantoni A, Legnini I, Capauto D, Rosa A, De Santis R et al (2017) FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat Commun 8:14741. https://doi.org/10.1038/ncomms14741
Acknowledgements
The authors thank all the patients and healthy controls that were willing to participate in the study, and Dr. Emanuela Boštjančič for her help with statistical analysis.
Funding
This work was supported by the Slovenian Research Agency (ARRS) under PhD thesis grant for young researcher Ana Dolinar and under research program P3-0054.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The study was approved by the National Medical Ethics Committee of Republic of Slovenia. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent was obtained from all individual participants included in the study.
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Dolinar, A., Koritnik, B., Glavač, D. et al. Circular RNAs as Potential Blood Biomarkers in Amyotrophic Lateral Sclerosis. Mol Neurobiol 56, 8052–8062 (2019). https://doi.org/10.1007/s12035-019-1627-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-019-1627-x