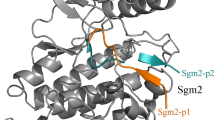
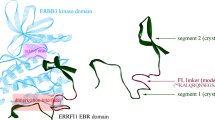
Abstract
The ErbB family of receptor tyrosine kinases (RTKs) contains four members: EGFR, ErbB2, ErbB3 and ErbB4; they are involved in the tumorigenesis of diverse cancers and can be inhibited natively by receptor-associated late transducer (RALT), a negative feedback regulator of ErbB signaling in human hepatocytes and hepatocellular carcinoma. Although the biological effects of RALT on EGFR kinase have been widely documented previously, the binding behavior of RALT to other ErbB/RTK kinases still remains largely unexplored. Here, the intermolecular interactions of RALT ErbB-binding region (EBR) as well as its functional sections and peptide segments with ErbBs and other human RTKs were systematically investigated at molecular and structural levels, from which we were able to identify those potential kinase targets of RALT protein, and to profile the affinity, specificity and cross-reactivity of RALT EBR domain and its sub-regions against various RTKs. It is revealed that RALT can target all the four ErbB kinases with high affinity for EGFR/ErbB2/ErbB4 and moderate affinity for ErbB3, but generally exhibits modest affinity to other RTKs, albeit few kinases such as LTK, EPHB6, MET and MUSK were also top-ranked as the unexpected targets of RALT. Peptide segments covering the key binding regions of RALT EBR domain were identified with computational alanine scanning, which were then optimized to obtain a number of designed peptide mutants with improved selectivity between different top-ranked RTKs.








Similar content being viewed by others
Availability of data and materials
The data that support the findings of this study are available upon request.
References
Anastasi S, Fiorentino L, Fiorini M, Fraioli R, Sala G, Castellani L, Alemà S, Alimandi M, Segatto O (2003) Feedback inhibition by RALT controls signal output by the ErbB network. Oncogene 22:4221–4234
Anastasi S, Baietti MF, Frosi Y, Alemà S, Segatto O (2007) The evolutionarily conserved EBR module of RALT/MIG6 mediates suppression of the EGFR catalytic activity. Oncogene 26:7833–7846
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The protein data bank. Nucleic Acids Res 28:235–242
Chen K, Huang L, Shen B (2019) Rational cyclization-based minimization of entropy penalty upon the binding of Nrf2-derived linear peptides to Keap1: a new strategy to improve therapeutic peptide activity against sepsis. Biophys Chem 244:22–28
Ciemny M, Kurcinski M, Kamel K, Kolinski A, Alam N, Schueler-Furman O, Kmiecik S (2018) Protein–peptide docking: opportunities and challenges. Drug Discov Today 23:1530–1537
Deng Y, Li J (2017) Rational optimization of tumor suppressor-derived inhibitor selectivity between oncogene tyrosine kinases ErbB1 and ErbB2. Arch Pharm 350:1700181
Du Z, Lovly CM (2018) Mechanisms of receptor tyrosine kinase activation in cancer. Mol Cancer 17:58
Ferby I, Reschke M, Kudlacek O, Knyazev P, Pantè G, Amann K, Sommergruber W, Kraut N, Ullrich A, Fässler R, Klein R (2006) Mig6 is a negative regulator of EGF receptor-mediated skin morphogenesis and tumor formation. Nat Med 12:568–573
Fiorentino L, Pertica C, Fiorini M, Talora C, Crescenzi M, Castellani L, Alemà S, Benedetti P, Segatto O (2000) Inhibition of ErbB-2 mitogenic and transforming activity by RALT, a mitogen-induced signal transducer which binds to the ErbB-2 kinase domain. Mol Cell Biol 20:7735–7750
Furge KA, Zhang YW, Vande Woude GF (2000) Met receptor tyrosine kinase: enhanced signaling through adapter proteins. Oncogene 19:5582–5589
Gordon JC, Myers JB, Folta T, Shoja V, Heath LS, Onufriev A (2005) H++: a server for estimating pKas and adding missing hydrogens to macromolecules. Nucleic Acids Res 33:W368–W371
Hackel PO, Gishizky M, Ullrich A (2001) Mig-6 is a negative regulator of the epidermal growth factor receptor signal. Biol Chem 382:1649–1662
Homeyer N, Horn AH, Lanig H, Sticht H (2016) AMBER force-field parameters for phosphorylated amino acids in different protonation states: phosphoserine, phosphothreonine, phosphotyrosine, and phosphohistidine. J Mol Model 12:281–289
Hubbard SR (1999) Structural analysis of receptor tyrosine kinases. Prog Biophys Mol Biol 71:343–358
Johansson MU, Zoete V, Michielin O, Guex N (2012) Defining and searching for structural motifs using DeepView/Swiss-PdbViewer. BMC Bioinform 13:173
Ko J, Park H, Heo L, Seok C (2012) GalaxyWEB server for protein structure prediction and refinement. Nucleic Acids Res 40:W294–W297
Kortemme T, Kim DE, Baker D (2004) Computational alanine scanning of protein-protein interfaces. Sci STKE 2014:pl2
Krivov GG, Shapovalov MV, Dunbrack RL (2009) Improved prediction of protein side-chain conformations with SCWRL4. Proteins 77:778–795
Ledda F, Paratcha G (2007) Negative regulation of receptor tyrosine kinase (RTK) signaling: a developing field. Biomark Insights 2:45–58
Li N, Wei M (2017) Conversion of Mig6 peptide from the nonbinder to binder of lung cancer-related EGFR by phosphorylation and cyclization. Artif Cells Nanomed Biotechnol 45:1023–1028
Li Z, Miao Q, Yan F, Meng Y, Zhou P (2019) Machine learning in quantitative protein–peptide affinity prediction: implications for therapeutic peptide design. Curr Drug Metab 20:170–176
London N, Movshovitz-Attias D, Schueler-Furman O (2010) The structural basis of peptide–protein binding strategies. Structure 18:188–199
Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S (2002) The protein kinase complement of the human genome. Science 298:1912–1934
McClendon CL, Kornev AP, Gilson MK, Taylor SS (2014) Dynamic architecture of a protein kinase. Proc Natl Acad Sci USA 111:E4623–E4631
Ouyang Y, Zhao L, Zhang Z (2018) Characterization of the structural ensembles of p53 TAD2 by molecular dynamics simulations with different force fields. Phys Chem Chem Phys 20:8676–8684
Park E, Kim N, Ficarro SB, Zhang Y, Lee BI, Cho A, Kim K, Park AKJ, Park WY, Murray B, Meyerson M, Beroukhim R, Marto JA, Cho J, Eck MJ (2015) Structure and mechanism of activity-based inhibition of the EGF receptor by Mig6. Nat Struct Mol Biol 2:703–711
Parker KC, Bednarek MA, Coligan JE (1994) Scheme for ranking potential HLA-A2 binding peptides based on independent binding of individual peptide side-chains. J Immunol 152(1):163–175
Qiao Z, Wang S (2021) Directed molecular engineering of Mig6 selectivity between proto-oncogene ErbB family receptor tyrosine kinases. Biotech Bioprocess Eng 26:277–285
Reschke M, Ferby I, Stepniak E, Seitzer N, Horst D, Wagner EF, Ullrich A (2010) Mitogen-inducible gene-6 is a negative regulator of epidermal growth factor receptor signaling in hepatocytes and human hepatocellular carcinoma. Hepatology 51:1383–1390
Robinson DR, Wu YM, Lin SF (2000) The protein tyrosine kinase family of the human genome. Oncogene 19:5548–5557
Roskoski R (2015) A historical overview of protein kinases and their targeted small molecule inhibitors. Pharmacol Res 100:1–23
Seifert E (2014) OriginPro 9.1: scientific data analysis and graphing software-software review. J Chem Inf Model 54:1552
UniProt C (2017) UniProt: the universal protein knowledgebase. Nucleic Acids Res 45:D158–D169
Vanhee P, Stricher F, Baeten L, Verschueren E, Lenaerts T, Serrano L, Rousseau F, Schymkowitz J (2009) Protein–peptide interactions adopt the same structural motifs as monomeric protein folds. Structure 17:1128–1136
Wang Z (2017) ErbB receptors and cancer. Methods Mol Biol 1652:3–35
Wang Z, Longo PA, Tarrant MK, Kim K, Head S, Leahy DJ, Cole PA (2011) Mechanistic insights into the activation of oncogenic forms of EGF receptor. Nat Struct Mol Biol 18:1388–1393
Wang Z, Raines LL, Hooy RM, Roberson H, Leahy DJ, Cole PA (2013) Tyrosine phosphorylation of mig6 reduces its inhibition of the epidermal growth factor receptor. ACS Chem Biol 8:2372–2376
Wang W, Ye W, Jiang C, Luo R, Chen HF (2014) New force field on modeling intrinsically disordered proteins. Chem Biol Drug Des 84:253–269
Wang H, Rao B, Lou J, Li J, Liu Z, Li A, Cui G, Ren Z, Yu Z (2020) The function of the HGF/c-Met axis in hepatocellular carcinoma. Front Cell Dev Biol 8:55
Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T (2018) SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46:W296–W303
You H, Ding W, Dang H, Jiang Y, Rountree CB (2011) c-Met represents a potential therapeutic target for personalized treatment in hepatocellular carcinoma. Hepatology 54:879–889
Yu H, Zhou P, Deng M, Shang Z (2014) Indirect readout in protein–peptide recognition: a different story from classical biomolecular recognition. J Chem Inf Model 54:2022–2032
Yu XD, Guo AF, Zheng GH, Yang XW, Shi PC (2016) Design and optimization of peptide ligands to target breast cancer-positive Her2 by grafting and truncation of Mig6 peptide. Int J Pept Res Ther 22:229–236
Zhang X, Pickin KA, Bose R, Jura N, Cole PA, Kuriyan J (2007) Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface. Nature 450:741–744
Zhang D, He D, Huang L, Xu Y, Liu L (2018) Rational design and cyclization of Mig6 peptide to restore its binding affinity for ErbB family receptor tyrosine kinases. Int J Pept Res Ther 24:71–76
Zhang Q, Jing T, Cui X, Zhao L (2021) Rational molecular profiling of receptor-associated late transducer peptide selectivity across Her/Rtk kinases. Int J Pept Res Ther 27:1945–1951
Zhong H, He J, Yu J, Li X, Mei Y, Hao L, Wu X (2021) Mig6 not only inhibits EGFR and HER2 but also targets HER3 and HER4 in a differential specificity: implications for targeted esophageal cancer therapy. Biochimie 190:132–142
Zhou P, Miao Q, Yan F, Li Z, Jiang Q, Wen L, Meng Y (2019) Is protein context responsible for peptide-mediated interactions? Mol Omics 15:280–295
Zhou P, Yan F, Miao Q, Chen Z, Wang H (2021a) Why the first self-binding peptide of human c-Src kinase does not contain class II motif but can bind to its cognate Src homology 3 domain in class II mode? J Biomol Struct Dyn 39:310–318
Zhou P, Liu Q, Wu T, Miao Q, Shang S, Wang H, Chen Z, Wang S, Wang H (2021b) Systematic comparison and comprehensive evaluation of 80 amino acid descriptors in peptide QSAR modeling. J Chem Inf Model 61:1718–1731
Zhuo ZH, Sun YZ, Jin PN, Li FY, Zhang YL, Wang HL (2016) Selective targeting of MAPK family kinases JNK over p38 by rationally designed peptides as potential therapeutics for neurological disorders and epilepsy. Mol Biosyst 12:2532–2540
Acknowledgements
This work was supported by the LPH foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Research involving human participants and/or animals
Not applicable.
Informed consent
Not applicable.
Additional information
Handling editor: F. Polticelli.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Lu, G., Li, X., Zhang, J. et al. Molecular insight into the affinity, specificity and cross-reactivity of systematic hepatocellular carcinoma RALT interaction profile with human receptor tyrosine kinases. Amino Acids 53, 1715–1728 (2021). https://doi.org/10.1007/s00726-021-03083-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00726-021-03083-8