Abstract
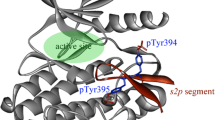
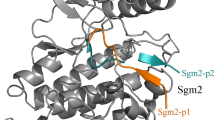
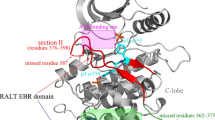
The ErbB signaling pathway plays important roles in normal physiology and cancer, which consists of four proto-oncogene receptor tyrosine kinases ErbB1/EGFR ErbB2/Her2, ErbB3/Her3, and ErbB4/Her4. Selective targeting of different ErbB kinases would result in distinct therapeutic effects, but traditional small-molecule inhibitors generally exhibit a strong cross-reactivity over these kinases due to the very high conservation in kinase’s active site. Instead of developing small-molecule drugs to selectively target the conserved active site of ErbB kinases, we herein attempt to design peptide agents for selectively disrupting the dimerization event of these kinases at their asymmetric dimer interfaces that have a relatively low homology. Three hotspot peptides S1P1, S1P2, and S1P3 are split from the functional segment 1 (Seg1) of mitogen-inducible gene 6 (Mig6), a natural EGFR-inhibitory protein that has been observed to inactivate the kinase by disrupting its dimerization. We demonstrate that the Mig6 peptides not only inhibit EGFR but also bind Her2, Her3, and Her4, although the peptide affinities to the four ErbB kinases are different considerably, exhibiting a typical selectivity. The S1P2 peptide locates in the core binding region of Mig6 Seg1 and contributes significantly to the segment interaction with kinases. An iteration algorithm is employed to guide the directed molecular engineering of S1P2 peptide selectivity towards each of the four kinases. Hundreds of parallel evolution running yield a series of peptide candidates with potential selectivity, which are then substantiated by fluorescence-based assays. The designed EGFR-selective peptide S1P2-p1EGFR is determined to have a moderate affinity to EGFR (Kd = 56 µM) and a high selectivity for EGFR over Her2, Her3, and H4 (FEGFR = 10.1-fold), which is improved considerably relative to wild-type S1P2 peptide (FEGFR = 2.7-fold). Structural examination observes different noncovalent interaction modes at the complex interfaces of S1P2-p1EGFR with EGFR and other three kinases, revealing a molecular origin of the peptide selectivity.
Similar content being viewed by others
References
Lemmon, M. A. and J. Schlessinger (2010) Cell signaling by receptor tyrosine kinases. Cell. 141: 1117–1134.
Wieduwilt, M. J. and M. M. Moasser (2008) The epidermal growth factor receptor family: biology driving targeted therapeutics. Cell. Mol. Life Sci. 65: 1566–1584.
Roskoski, R. (2014) The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacol. Res. 79: 34–74.
Walker, F., S. G. Orchard, R. N. Jorissen, N. E. Hall, H. H. Zhang, P. A. Hoyne, T. E. Adams, T. G. Johns, C. Ward, T. P. J. Garrett, H. J. Zhu, M. Nerrie, A. M. Scott, E. C. Nice, and A. W. Burgess (2004) CR1/CR2 interactions modulate the functions of the cell surface epidermal growth factor receptor. J. Biol. Chem. 279: 22387–22398.
Singh, D., B. K. Attri, R. K. Gill, and J. Bariwal (2016) Review on EGFR inhibitors: critical updates. Mini. Rev. Med. Chem. 16: 1134–1166.
Dawson, J. P., M. B. Berger, C. C. Lin, J. Schlessinger, M. A. Lemmon, and K. M. Ferguson (2005) Epidermal growth factor receptor dimerization and activation require ligand-induced conformational changes in the dimer interface. Mol. Cell. Biol. 25: 7734–7742.
Hackel, P. O., M. Gishizky, and A. Ullrich (2001) Mig-6 is a negative regulator of the epidermal growth factor receptor signal. Biol. Chem. 382: 1649–1662.
Zhang, X., K. A. Pickin, R. Bose, N. Jura, P. A. Cole, and J. Kuriyan (2007) Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface. Nature. 450: 741–744.
Ferby, I., M. Reschke, O. Kudlacek, P. Knyazev, G. Pantè, K. Amann, W. Sommergruber, N. Kraut, A. Ullrich, R. Fässler, and R. Klein (2006) Mig6 is a negative regulator of EGF receptor-mediated skin morphogenesis and tumor formation. Nat. Med. 12: 568–573.
Wang, Z., L. L. Raines, R. M. Hooy, H. Roberson, D. J. Leahy, and P. A. Cole (2013) Tyrosine phosphorylation of mig6 reduces its inhibition of the epidermal growth factor receptor. ACS Chem. Biol. 8: 2372–2376.
Yu, X. D., R. Yang, and C. J. Leng (2016) Truncation, modification, and optimization of MIG6(segment 2) peptide to target lung cancer-related EGFR. Comput. Biol. Chem. 61: 251–257.
Li, N. and M. Wei (2017) Conversion of MIG6 peptide from the nonbinder to binder of lung cancer-related EGFR by phosphorylation and cyclization. Artif. Cells Nanomed. Biotechnol. 45: 1023–1028.
Wang, H., C. Guo, D. Ren, T. Xu, Y. Cao, W. Xiao, and W. Jiao (2017) Let it bind: cyclization of Mig-6 segment 2 to target EGFR signaling in lung cancer. Med. Chem. Res. 26: 1747–1752.
Yu, X. D., A. F. Guo, G. H. Zheng, X. W. Yang, and P. C. Shi (2016) Design and optimization of peptide ligands to target breast cancer-positive HER2 by grafting and truncation of MIG6 peptide. Int. J. Pept. Res. Ther. 22: 229–236.
Zhuo, Z. H., Y. Z. Sun, P. N. Jin, F. Y. Li, Y. L. Zhang, and H. L. Wang (2016) Selective targeting of MAPK family kinases JNK over p38 by rationally designed peptides as potential therapeutics for neurological disorders and epilepsy. Mol. Biosyst. 12: 2532–2540.
Gu, Z., T. Yan, and F. Yan (2020) Rational design and improvement of the dimerization-disrupting peptide selectivity between ROCK-I and ROCK-II kinase isoforms in cerebrovascular diseases. J. Mol. Recognit. 33: e2835.
Subrizi, A., E. Tuominen, A. Bunker, T. Róg, M. Antopolsky, and A. Urtti (2012) Tat(48–60) peptide amino acid sequence is not unique in its cell penetrating properties and cell-surface glycosaminoglycans inhibit its cellular uptake. J. Control Release. 158: 277–285.
Park, E., N. Kim, S. B. Ficarro, Y. Zhang, B. I. Lee, A. Cho, K. Kim, A. K. J. Park, W. Y. Park, B. Murray, M. Meyerson, R. Beroukhim, J. A. Marto, J. Cho, and M. J. Eck (2015) Structure and mechanism of activity-based inhibition of the EGF receptor by Mig6. Nat. Struct. Mol. Biol. 22: 703–711.
Berman, H. M., J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov, and P. E. Bourne (2000) The protein data bank. Nucleic Acids Res. 28: 235–242.
Zhou, P., S. Hou, Z. Bai, Z. Li, H. Wang, Z. Chen, and Y. Meng (2018) Disrupting the intramolecular interaction between proto-oncogene c-Src SH3 domain and its self-binding peptide PPII with rationally designed peptide ligands. Artif. Cells Nanomed. Biotechnol. 46: 1122–1131.
Guex, N. and M. C. Peitsch (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 18: 2714–2723.
Krivov, G. G., M. V. Shapovalov, and R. L. Dunbrack Jr (2009) Improved prediction of protein side-chain conformations with SCWRL4. Proteins. 77: 778–795.
Ko, J., H. Park, L. Heo, and C. Seok (2012) GalaxyWEB server for protein structure prediction and refinement. Nucleic Acids Res. 40: W294–W297.
Gordon, J. C., J. B. Myers, T. Folta, V. Shoja, L. S. Heath, and A. Onufriev (2005) H++: a server for estimating pKas and adding missing hydrogens to macromolecules. Nucleic Acids Res. 33: W368–W371.
Maier, J. A., C. Martinez, K. Kasavajhala, L. Wickstrom, K. E. Hauser, and C. Simmerling (2015) ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 11: 3696–3713.
Yang, C., S. Zhang, P. He, C. Wang, J. Huang, and P. Zhou (2015) Self-binding peptides: folding or binding. J. Chem. Inf. Model. 55: 329–342.
Yang, C., S. Zhang, Z. Bai, S. Hou, D. Wu, J. Huang, and P. Zhou (2016) A two-step binding mechanism for the self-binding peptide recognition of target domains. Mol. Biosyst. 12: 1201–1213.
Ndagi, U., N. N. Mhlongo, and M. E. Soliman (2018) Emergence of a promising lead compound in the treatment of triple negative breast cancer: an insight into conformational features and ligand binding landscape of c-Src protein with UM-164. Appl. Biochem. Biotechnol. 185: 655–675.
Yang, C., C. Wang, S. Zhang, J. Huang, and P. Zhou (2015) Structural and energetic insights into the intermolecular interaction among human leukocyte antigens, clinical hypersensitive drugs and antigenic peptides. Mol. Simul. 41: 741–751.
Zhou, P., F. Yan, Q. Miao, Z. Chen, and H. Wang (2021) Why the first self-binding peptide of human c-Src kinase does not contain class II motif but can bind to its cognate Src homology 3 domain in class II mode? J. Biomol. Struct. Dyn. 39: 310–318.
Berendsen, H. J. C., J. P. M. Postma, W. F. Van Gunsteren, A. Dinola, and J. R. Haak (1984) Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81: 3684.
Gonnet, P (2007) P-SHAKE: a quadratically convergent SHAKE in O(n2). J. Comput. Phys. 220: 740–750.
Salomon-Ferrer, R., A. W. Götz, D. Poole, S. Le Grand, and R. C. Walker (2013) Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh Ewald. J. Chem. Theory Comput. 9: 3878–3888.
Le Grand, S., A. W. Götz, and R. C. Walker (2013) SPFP: Speed without compromise—a mixed precision model for GPU accelerated molecular dynamics simulations. Comput. Phys. Commun. 184: 374–380.
Genheden, S. and U. Ryde (2015) The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert. Opin. Drug Discov. 10: 449–461.
Bai, Z., S. Hou, S. Zhang, Z. Li, and P. Zhou (2017) Targeting self-binding peptides as a novel strategy to regulate protein activity and function: a case study on the proto-oncogene tyrosine protein kinase c-Src. J. Chem. Inf. Model. 57: 835–845.
Weng, G., E. Wang, F. Chen, H. Sun, Z. Wang, T. Hou (2019) Assessing the performance of MM/PBSA and MM/GBSA methods. 9. Prediction reliability of binding affinities and binding poses for protein-peptide complexes. Phys. Chem. Chem. Phys. 21: 10135–10145.
Li, Z., F. Yan, Q. Miao, Y. Meng, L. Wen, Q. Jiang, and P. Zhou (2019) Self-binding peptides: binding-upon-folding versus folding-upon-binding. J. Theor. Biol. 469: 25–34.
Bahar, I., T. R. Lezon, A. Bakan, and I. H. Shrivastava (2010) Normal mode analysis of biomolecular structures: functional mechanisms of membrane proteins. Chem. Rev. 110: 1463–1497.
Han, K. Q., G. Wu, and F. Lv (2013) Development of QSAR-improved statistical potential for the structure-based analysis of protein-peptide binding affinities. Mol. Inform. 32: 783–792.
Li, Z., Q. Miao, F. Yan, Y. Meng, and P. Zhou (2019) Machine learning in quantitative protein-peptide affinity prediction: implications for therapeutic peptide design. Curr. Drug Metab. 20: 170–176.
Kortemme, T., D. E. Kim, and D. Baker (2004) Computational alanine scanning of protein-protein interfaces. Sci. STKE. 2004: pl2.
Zhou, K., J. Lu, X. Yin, H. Xu, L. Li, and B. Ma (2019) Structure-based derivation and intramolecular cyclization of peptide inhibitors from PD-1/PD-L1 complex interface as immune checkpoint blockade for breast cancer immunotherapy. Biophys. Chem. 253: 106213.
Zhu, Z., C. Zhang, and W. Song (2017) Rational derivation, extension, and cyclization of self-inhibitory peptides to target TGF-β/BMP signaling in ONFH. Amino Acids. 49: 283–290.
Song, W., K. Wang, W. Wang, P. Yang, and X. Dang (2019) Grafting, stripping and stapling of helical peptides from the dimerization interface of ONFH-related bone morphogenetic protein-2. Protein J. 38: 12–22.
Deng, Y. and J. Li (2017) Rational optimization of tumor suppressor-derived peptide inhibitor selectivity between oncogene tyrosine kinases ErbB1 and ErbB2. Arch Pharm Chem Life Sci. 2017: 1700181.
Zhou, P., Q. Miao, F. Yan, Z. Li, Q. Jiang, L. Wen, and Y. Meng (2019) Is protein context responsible for peptide-mediated interactions? Mol. Omics. 15: 280–295.
Acknowledgements
All the authors declare that there are no conflicts of interest. The authors declare no conflict of interest.
Neither ethical approval nor informed consent was required for this study.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Qiao, Z., Wang, S. Directed Molecular Engineering of Mig6 Peptide Selectivity between Proto-oncogene ErbB Family Receptor Tyrosine Kinases. Biotechnol Bioproc E 26, 277–285 (2021). https://doi.org/10.1007/s12257-020-0102-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12257-020-0102-x