Abstract
Receptor tyrosine kinases (RTKs) play an important role in a variety of cellular processes including growth, motility, differentiation, and metabolism. As such, dysregulation of RTK signaling leads to an assortment of human diseases, most notably, cancers. Recent large-scale genomic studies have revealed the presence of various alterations in the genes encoding RTKs such as EGFR, HER2/ErbB2, and MET, amongst many others. Abnormal RTK activation in human cancers is mediated by four principal mechanisms: gain-of-function mutations, genomic amplification, chromosomal rearrangements, and / or autocrine activation. In this manuscript, we review the processes whereby RTKs are activated under normal physiological conditions and discuss several mechanisms whereby RTKs can be aberrantly activated in human cancers. Understanding of these mechanisms has important implications for selection of anti-cancer therapies.
Similar content being viewed by others
Background
Receptor tyrosine kinases (RTKs) are a subclass of tyrosine kinases that are involved in mediating cell-to-cell communication and controlling a wide range of complex biological functions, including cell growth, motility, differentiation, and metabolism. There are 58 known RTKs in humans [1, 2], and all RTKs share a similar protein structure comprised of an extracellular ligand binding domain, a single transmembrane helix, and an intracellular region that contains a juxtamembrane regulatory region, a tyrosine kinase domain (TKD) and a carboxyl (C-) terminal tail [3]. Dysregulation of RTK signaling leads to many human diseases, especially cancer. Given the advent of the genomic era and the implementation of next generation sequencing (NGS) in cancer research as well as routine clinical practice, mutational landscapes have been established in almost all types of human tumors [4]. These genomic studies have revealed the presence of several different types of alterations in the genes encoding RTKs such as EGFR, HER2/ErbB2, MET, amongst many others. The presence of recurrent RTK genomic alterations raises the question about how they function in cancer development and how to best treat cancer patients whose tumors harbor certain RTK mutations. In this manuscript, we review the processes whereby RTKs are activated under normal physiological conditions and discuss several mechanisms whereby RTKs can be aberrantly activated in human cancers, which have important implications for selection of anti-cancer therapies.
Mechanisms of RTK activation under normal physiologic conditions
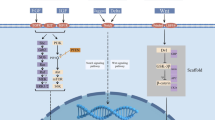
RTKs are generally activated by receptor-specific ligands. Growth factor ligands bind to extracellular regions of RTKs, and the receptor is activated by ligand-induced receptor dimerization and/or oligomerization [5] (Fig. 1a). For most RTKs, the resultant conformational changes enable trans-autophosphorylation of each TKD and release of the cis-autoinhibition [6]. This conformational change allows the TKD to assume an active conformation. Autophosphorylation of RTKs also recruits and activates a wide variety of downstream signaling proteins which contain Src homology-2 (SH2) or phosphotyrosine-binding (PTB) domains. These domains bind to specific phosphotyrosine residues within the receptor and engage downstream mediators that propagate critical cellular signaling pathways [7].
Mechanisms of physiological and oncogenic RTK activation. a Schematic representation of RTK activation in normal physiology. RTKs are activated through formation of inter-molecular dimerization in the presence of ligands, resulting in kinase activation and phosphorylation of the receptor C-terminal tail. b Schematic representation of potential gain-of-function mutations in the various subdomains of an RTK. The mutations lead to constitutive activation of the RTK, typically in the absence of ligand. c Overexpression of RTKs – often as a result of genomic amplification of the RTK gene - leads to increased local concentration of receptors
Ligand-induced dimerization of RTK extracellular regions
In general, there are four modes of RTK dimerization which lead to activation of the tyrosine kinase domain. In the first mode, receptor dimerization is completely ligand mediated without any direct contact between the extracellular regions of the two receptors, such as in the case of TrkA (NGF receptor) [8]. In the second mode, dimerization is instead completely receptor mediated without any physical interaction between two activating ligands, as in the case of ErbB family members (EGFR, HER2/ErbB2, HER3/ErbB3, and HER4/ErbB4) [9]. In the third mode, ligand homodimers bind to two receptor molecules, which then interact with each other across the dimer interface, such as the case for KIT (SCF receptor) [10]. In the fourth mode, in addition to a combination of bivalent ligand binding and direct receptor-receptor contacts, accessory molecules also participate in receptor dimerization. For example, the FGFR family of RTKs uses heparin or heparan sulfate as accessory molecules in this mode [11, 12].
Notably, a subset of RTKs forms dimers or high-order oligomers even without activating ligands. The receptors stay in dynamic equilibrium between monomers and dimers. For EGFR and many other RTKs, monomers predominate before ligand-binding [13]. For the insulin receptor (IR), dimers predominate even without ligands [14, 15]. The pre-formed dimers exist as either “inactive” or “active” form. The “inactive” dimers are likely in dynamic equilibrium with “active” dimers. An active dimer will be stabilized by ligand binding, whereas an inactive dimer will be activated by ligand binding through conformational changes. In both scenarios, the ligand binding will shift the equilibrium to the formation of ligand-induced dimerization [13,14,15].
The ErbB family is of particular interest in cancer biology, and therefore discussed here in additional detail. The extracellular regions of the ErbB receptors family include four subdomains (I-IV) [16]. In the absence of ligands, the intracellular TKD is inactive, and the extracellular region adopts a “tethered” configuration in which the dimerization arm (a β-hairpin within subdomain II of the ECD) is entirely buried by intra-molecular interactions with domain IV and forms intra-molecular autoinhibitory interactions. Ligand simultaneously binds to two sites (subdomain I and subdomain III) within the extracellular region of one receptor, rather than spanning two separate receptors as seen for NGF [8], SCF [10], or FGF receptor [17]. Ligand binding induces a dramatic conformational change that “extends” the extracellular region and exposes the previously buried dimerization arm to an active conformation. With the dimerization arm exposed, the extracellular region of the receptor dimerizes [18], inducing intracellular conformational changes so that they can enable kinase activation [9].
Activation of intracellular tyrosine kinase domains
Numerous studies have been performed to determine how physiological information is transmitted from the exterior to the interior of the cells. Before activation, the TKD is in a condition of cis-autoinhibition by certain intra-molecular interactions unique for each receptor [19, 20]. Ligand-induced dimerization releases this cis-autoinhibition. FGFR, IR, and IGF-1R receptors are autoinhibited by the activation loop, which directly contacts the active site of the kinase and disrupts ATP and substrate binding [21, 22]. KIT and Eph receptors are regulated by juxtamembrane autoinhibition, in which the juxtamembrane region interacts with components within the active site of the kinase—thereby stabilizing an inactive state [20, 23]. For the TEK, MET, and RON (MST-1R) receptors, the C-terminal tail contacts the active site of the TKD, thus inhibiting substrate access [19]. This interaction stabilizes an inactive conformation which exerts a strong autoinhibition on kinase activity. Ligand-induced dimerization induces trans-phosphorylation of key tyrosine residues, resulting in destabilization of these autoinhibitory interactions and therefore, allowing the kinase to assume an active conformation.
Again, calling out the unique properties of the ErbB family of RTKs – the kinase activity of these receptors is activated through a unique allosteric mechanism whereby the C-lobe of one kinase domain in the dimer pair (the so called ‘activator’ kinase) physically contacts the N-lobe of the other kinase domain in the dimer pair (the so called ‘receiver’ kinase). This physical interaction induces conformational changes in the N-lobe of receiver kinase [9] which induces activation of the ‘receiver’ kinase domain and trans-phosphorylation of tyrosine residues in the C-terminal tail of the ‘activator’. Phosphorylation of the activation loop is not involved in this mechanism [24, 25].
Mechanism of activation of downstream signaling
Activation and subsequent autophosphorylation of RTKs results in the recruitment of a wide range of downstream signaling proteins. Most autophosphorylation sites function as binding sites for SH2 or PTB domain containing signaling proteins. SH2 domain containing proteins can be recruited directly to the receptor, or indirectly to the receptor through docking proteins which bind to RTKs via their PTB domains. Docking proteins function as “assembly platforms” to recruit additional signaling molecules containing SH2 or other domains [5, 26]. The presence of several phosphotyrosines and the involvement of various docking proteins confer activated RTKs the ability to recruit and regulate a wide range of signaling pathways including RAS/MAPK, PI-3 K/AKT, and JAK2/STAT signaling. Therefore, RTKs function as a node which transfers complicated information regarding cell growth and migration from the extracellular milieu ultimately to the cell nucleus to activate transcriptional pathways involved in regulating many cellular processes.
Summary of RTK activation under normal physiologic conditions
Several decades of intricate structural and biochemical studies have revealed the complicated mechanisms whereby RTKs are activated in a ligand mediated way to propagate cellular signals. A detailed understanding of receptor physiology is crucial to fully understand how and why oncogenic mutations in RTKs disrupt this normal biology, resulting in a dysregulation of cell growth, aberrant cell signaling, and altered metabolism in tumor cells.
Oncogenic activation of receptor tyrosine kinases
Under normal physiologic conditions, the level of RTK activity is tightly balanced by the mechanisms described above and by additional molecules, including tyrosine phosphatases [27]. RTKs acquire transforming abilities through several mechanisms, and the final consequence is the disruption of the balance between cell growth/proliferation and cell death [5]. When temporal and spatial regulation are taken into consideration, dysregulated RTK signaling becomes even more complicated [28]. Constitutive activation may confer oncogenic properties upon normal cells and trigger RTK-induced oncogenesis [29]. Four principal mechanisms lead to constitutive RTK activation in human cancers: gain-of-function mutations, genomic amplification, chromosomal rearrangements, and / or autocrine activation [6]. Here, we discuss these four oncogenic activating mechanisms including a special intragenic duplication – kinase domain duplication (KDD).
Activation by gain-of-function mutations
A gain-of-function mutation in an RTK leads to aberrant downstream signal transduction, not subjected to the normal ‘checks and balances’ that occur with physiological signaling. Of particular interest is the identification and functional characterization of ‘driver mutations’ - defined as mutations that can confer a selective growth advantage to the cells [4]. These ‘driver mutations’ can shed light on the understanding of cancer initiation and progression and can also provide potential opportunities for targeted treatments. Somatic mutations in the genes encoding RTKs typically cluster in evolutionally conserved residues, such as the DFG motif in the kinase activation loop and around the nucleotide-binding pocket. These conserved residues (D, F, and G) play key roles in ATP binding and catalytic activity [30, 31].
Somatic EGFR mutations serve as excellent examples to illustrate the mutational spectrum of RTKs. The entire EGFR TKD is encoded by exons 18–24. EGFR mutations predominantly cluster in exons 18–21, which are adjacent to the ATP-binding pocket [32]. Approximately 90% of these mutations are small in-frame deletions within exon 19 or L858R point mutation within exon 21 [33,34,35]. These mutations hyperactivate the kinase and, subsequently, its downstream signaling, conferring oncogenic properties [32, 36, 37]. Numerous large international clinical trials have now shown that patients whose tumors harbor activating somatic EGFR TKD mutations are uniquely sensitive to treatment with EGFR tyrosine kinase inhibitors (TKIs) [38,39,40,41,42,43,44,45].
Mutations can also occur in extracellular domain (ECD), transmembrane domain (TMD) and juxtamembrane domain (JMD) of RTKs. Three missense mutations within the EGFR ECD (P596L, G598 V, and A289V) were previously reported in glioblastoma (GBM) [46, 47]. These mutations are associated with increased expression of EGFR protein, which undergoes phosphorylation in the absence of ligand stimulation [46]. In contrast to lung cancer patients with EGFR TKD mutations, GBM patients with EGFR ECD mutations have shown disappointing clinical outcomes when treated with the EGFR TKIs, erlotinib and gefitinib [48, 49]. Studies suggest that the EGFR ECD mutations adopt the inactive conformation (compared to EGFR TKD mutations which adopt the active conformation), and the net effect is that EGFR ECD mutations may be better inhibited with EGFR targeted therapies that bind to the inactive form of the receptor [50]. Point mutations in the FGFR3 ECD (specifically, S249C) were reported in carcinomas of the uterine cervix [51]. These mutations result in unpaired cysteine residues, allowing abnormal receptor dimerization through intermolecular disulfide bonding [52]. Mutations within ECD of other RTKs have also been reported, including RET in thyroid cancer [53] and KIT in gastrointestinal stromal tumor (GIST) [54]. HER2 G660D and V659E mutations within the TMD act as driver mutations in non-small cell lung cancer (NSCLC) [55]. HER2 V659 mutations are also found in a patient with Li-Fraumeni syndrome [56]. These mutations disrupt specific protein-protein and protein-lipid interactions within the HER2 TMD that are essential for proper receptor dimerization [57]. It has been also shown that these two TMD mutations exhibit lower protein turnover than wild-type HER2 [58]. In in vitro models, HER2 V659E exhibits sensitivity to two TKIs - lapatinib [56] and afatinib [59], indicating TMD mutations could serve as actionable therapeutic targets. Finally, mutations within the JMD release autoinhibitory juxtamembrane interactions and subsequently hyperactivate these RTKs, such as KIT V560G and PDGFRA V561D mutation in GIST [54]. Therefore, mutations within the ECD, TMD and JM of RTKs adopt alternative activating mechanisms compared to mutations within the TKD. It has been observed that patients with GIST harboring mutations within the ECD, TMD, and/or JMD have different treatment response from TKD mutations to targeted therapy by using imatinib [54], a competitive inhibitor of KIT [60] and PDGFRA [61]. Gain-of-function mutations in the various subdomains of the RTKs described above are represented schematically in Fig. 1b.
Overexpression and genomic amplification
Overexpression of RTKs has been found in a variety of human cancers: EGFR in GBM [62], lung [63], esophageal [64] and thyroid cancer [65]; HER2/ErbB2 in lung [66], bladder [67], breast [68] and gastric cancer [69, 70]; and MET in lung [71] and gastric cancer [72]. Overexpression leads to increased local concentration of receptor, which results in elevated RTK signaling and overwhelms the antagonizing regulatory effects [73]. While gene amplification is the major mechanism which leads to overexpression of RTKs, additional mechanisms of RTK overexpression include transcriptional/translational enhancement [74, 75], oncogenic viruses [64], derailment of normal regulatory mechanisms such as loss of phosphatases [76] or other negative regulators [77, 78]. Regardless of mechanism, overexpression of RTKs has been associated with poor outcomes in some cancer patients, such as EGFR and HER3 in breast cancer [79].
Gene amplification is characterized by a process that increases the copy number of a specific region of the genome [80]. Genomic amplification can occur as extrachromosomal elements (double minutes), repeated units at a single locus or distributed throughout the genome (distributed insertions) [81]. Double minutes tend to result in high level amplification (> 25 copies) while distributed insertions tend to low level amplification (5 to 25 copies) [62]. Gene amplification may be influenced by common chromosomal fragile sites, defects in DNA replication, or telomere dysfunction [80]. Amplification of many RTKs occurs in a variety of human cancers, such as EGFR, ERBB2 and MET [80]. Other RTK amplifications have also been reported in human cancers, including FGFR1 in lung and breast cancer [82, 83], FGFR3 in breast and bladder cancer [84, 85], ERBB4 in breast and gastric cancer [86, 87], FLT3 in colon cancer [88], KIT in melanoma and GIST [89, 90], and PDGFRA in GBM [91]. Amplification patterns differ largely even in the same tumor type [62]. For example, a recent study in GBM indicated that 88% of cases with high-level EGFR genomic amplification showed EGFR protein overexpression by immunohistochemistry, in contrast to 36% of the cases with low-level EGFR amplification [62]. Lastly, RTK amplification can occur either in the context of a wild-type or mutated allele. For example, EGFR amplification was found to occur preferentially on the mutated allele in EGFR-mutant lung cancer [92]. RTK amplifications also act as an avenue for tumor cells to escape therapeutic treatment. For example, MET amplification and HER2 amplification can be detected in EGFR-mutant lung cancers that become resistant to EGFR tyrosine kinase inhibitor therapy [93]. RTK overexpression is represented schematically in Fig. 1c.
Chromosomal rearrangements
Genomic studies have identified numerous chromosomal rearrangements which lead to the formation of novel tyrosine kinase fusion oncoproteins [94,95,96]. The importance of identifying these chromosomal rearrangements and the ensuing tyrosine kinase fusion is underscored by that fact that these aberrant fusion proteins are often therapeutically targetable with small molecule inhibitors. The first tyrosine kinase fusion identified was BCR-ABL, which derived from translocation t(9,22) – the so called ‘Philadelphia Chromosome’ – which fuses the gene encoding the ABL1 tyrosine kinase on chromosome 9 to the BCR gene on chromosome 22, to form the BCR-ABL fusion oncoprotein [97]. BCR-ABL is characteristically found in patients with chronic myelogenous leukemia (CML) and in some patients with acute lymphoblastic leukemia [98, 99]. Notably, the first tyrosine kinase inhibitor developed and approved by the US Food and Drug Administration (FDA) – imatinib – targets the ABL kinase and has revolutionized the treatment of patients with CML [100, 101].
While BCR-ABL occurs exclusively in leukemia, many of the subsequently discovered tyrosine kinase fusions occur in multiple tumor types, including both liquid and solid malignancies. For example, the translocation t(2,5) fuses the gene encoding the ALK tyrosine kinase on chromosome 2 to the NPM gene on chromosome 5, to form the NPM-ALK fusion oncoprotein [102], which is found in approximately 50% of anaplastic large cell lymphoma (ALCL) [103]. Almost 30 years after the identification of the NPM-ALK fusion, similar ALK tyrosine kinase fusions have been found in other tumor types. Most notably, ALK rearrangements occur in approximately 3–7% of NSCLCs [104], approximately 50% of all inflammatory myofibroblastic tumors (IMTs) [105, 106], 10% of Spitzoid neoplasms [107], as well as small percentages in colon cancer [94, 108, 109], thyroid cancer [94, 110], and several other types of malignancies [94, 102, 111]. Likewise, oncogenic tyrosine kinase fusions involving ROS1 have been identified in ~ 1% of NSCLCs [112], as well as in IMTs, cholangiocarcinoma, and GBM [94, 113]. RET kinase fusions have been recurrently detected in NSCLC and thyroid cancers [94, 114, 115]. Last but certainly not least, fusion oncoproteins involving the TRKA, TRKB, and TRKC tyrosine kinases (which are encoded by NTRK1, NTRK2, and NTRK3, respectively) have been identified across nine tumor types, including sarcoma, melanoma, gliomas, thyroid, lung, colon, breast, head and neck cancers) [94]. The fusion proteins have been reported as potent actionable targets in adult and children with TRK fusion positive cancers [116]. Numerous other tyrosine kinase fusions have been described, including those that incorporate EGFR [94, 117], HER2 [118], MET [94, 107], PDGFRa [119], and PDGFRb [94, 106]. These findings suggest that fusion events may have some common underlying etiology in human tumors. Several risk factors have been considered to contribute to the gene fusion events, including exposure to ionizing radiation [120, 121], topoisomerase poisons [122] and oxidative stress [123], but the precise molecular mechanisms remain elusive.
Despite the diversity of tyrosine kinase fusions which have been described, the structure of the resultant fusion oncoproteins retains a remarkable similarity. Fusions may occur in either the N-terminal or the C-terminal of the RTK, with the TKD preserved in both cases (Fig. 2a). If the genomic breakpoint occurs downstream of the exons encoding the full kinase domain (with preservation of the ECD, TMD, and JMD), then the resultant fusion protein will function as a membrane-bound receptor, such as the case for the EGFR-RAD51 fusion protein [117]. If the genomic breakpoint occurs upstream of the exons encoding the full kinase domain (with loss of the ECD, TMD, and JMD), then the resultant fusion protein will not be membrane bound. Instead, such proteins typically localize to the cytoplasm, as is the case for the EML4-ALK fusion protein [124]. Another characteristic of kinase fusions is the occurrence of multiple fusion partners within the same disease [94, 106, 125]. For example, there are at least nine known ROS1 fusion partners found in NSCLC, including SLC34A2, CD47, TPM3, SDC4, EZR, LRIG3, FIG, KDELR2 and CCDC6 [94].
Mechanisms of oncogenic RTK activation. a Chromosomal rearrangements result in the formation of a hybrid fusion oncoprotein consisting partly of the RTK and partly of the fusion partner, a distinct protein (shown in the figure by the yellow oval). These RTK fusion proteins can be membrane bound (left side of the figure) or cytoplasmic (right side of the figure) depending on the location of the genomic breakpoint. In either case, the result is an activated kinase domain. b Duplication of the tyrosine kinase domain could possibly form an intra-molecular dimer in the absence of ligands, resulting in RTK activation. c Schematic representation of autocrine activation of RTK signaling. Increased local concentration of ligand activated the RTK, resulting in RTK dimerization, increased kinase activity, and phosphorylation of the receptor C-terminal tail
Although these partners can vary, they share three features. First, the regulatory unit of the fusion partner dictates the expression of the fusion, placing the tyrosine kinase oncoprotein under the endogenous promoter of the fusion partner [108, 126]. Second, most fusion partners contribute an oligomerization domain, which promotes ligand independent constitutive activation of the kinase [94, 127, 128]. The most common oligomerization domains found in the fusion partners are coiled-coil domains. For example, EML4-ALK, the most common ALK fusion detected in NSCLC, homodimerizes by virtue of a coiled-coil domain in EML4 [124]. Disruption of the coiled-coil domain abrogates the ability of EML4-ALK to transform cells [124]. Third, the fusion partner also determines subcellular localization of the fusion [129, 130], and this may have profound effects on the protein interactions that the fusion encounters, affecting activation, signaling, function, and degradation of the fusion. As such, RTK fusions can regulate similar cell signaling pathways as the ‘parental’ RTK from which they are derived (including RAS/MAPK, PI-3 K/AKT, and JAK2/STAT [106, 117]) and/or possibly even new pathways based on their altered cellular localization.
Chromosomal rearrangements of RTKs lead to chimeric fusion proteins, which contribute to oncogene addiction [106, 117]. Inhibiting RTK fusions with target specific TKIs has proven to be an effective therapeutic strategy across numerous types of RTK fusion driven cancers – including ALK in ALCL [131], IMT [132] and lung cancer [133], RET in lung and thyroid cancer [134,135,136,137], ROS1 in GBM [138], lung cancer [139], and IMT [106], EGFR in lung cancer [117], and NTRK in IMT [140], lung [141], kidney [141], colon [140, 141] and other types of cancer [141].
Constitutive activation by kinase domain duplication
Intragenic partial duplication is a type of chromosomal rearrangement that confers cancer cells the ability to acquire new protein isoforms [142]. Kinase domain duplications (KDDs) constitute one type of intragenic partial duplication, resulting in a novel mechanism for RTK activation in tumor cells. For example, oncogenic EGFR-KDD and BRAF-KDD have been reported in human cancers, along with their responses to the respective targeted therapies against EGFR and BRAF. Recently, our group reported that EGFR-KDD is recurrently found in NSCLC [143]. We also found that EGFR-KDD occurred in other types of human tumors, including gliomas, sarcoma and Wilms’ tumor [143]. BRAF-KDD has been reported in gliomas and advanced acinic cell tumor [144, 145]. BRAF is an intracellular serine/threonine kinase; however, we discuss here as demonstration of principle. Most recently, a group of investigators has analyzed clinical genomic data from 114,200 human tumors and found recurrent KDD alterations involving several kinases, including the ErbB family (EGFR, ERBB2 and ERBB4), FGFR family (FGFR1, FGFR2 and FGFR3), NTKR family (NTRK1 and NTRK2), PDGFR family (PDGFRA and PDGFRB), and other kinases (BRAF, RET, MET, ROS1, ALK and KIT) [146]. In brain tumors, KDD occurs most frequently within EGFR, BRAF, PDGFRA, and FGFR3. In extracranial tumors, KDD was frequently found in RET, MET and ALK genes [146]. Overall, the frequency of KDD alterations was 0.62% (598 total KDDs in 114,200 cases analyzed).
In nature, gene duplication is one method by which species introduce genetic novelty or redundancy, thereby allowing them to adapt to various environmental conditions [147]. It is possible that KDDs in tumor cells can be selected for in response to pressure exerted by cancer therapy. For example, BRAF-KDD was identified as a new mechanism of drug resistance in patients with melanoma after BRAF inhibitor treatment [142]. Identification of EGFR-KDD amplification in the post-treatment biopsy suggested that KDD is also involved in the acquired resistance of EGFR TKI, afatinib [143].
To date, the most well studied KDD is the EGFR-KDD [143]. In normal biology, the presence of EGF ligands activates wild-type EGF receptor through the formation of an asymmetric dimer between two receptor molecules [9]. Considering that EGFR-KDD contains two tandem, in-frame tyrosine kinase domains, it is possible that the mode of activation of the EGFR-KDD variant involves constitutive intra-molecular dimerization (Fig. 2b). Therefore, for this variant, EGFR signaling can be activated in a ligand independent manner. Preclinical modeling of the EGFR-KDD protein validated this potential activation mechanism in silico and in vitro. Notably, EGFR-KDD activation is quite distinct from the molecular mechanisms governing activation of EGFR kinase domain mutants described above (e.g., L858R, exon 19 deletion), underscoring the importance of considering how genomic findings alter protein structure and function to result in an oncogenic variant.
With respect to BRAF-KDD, most of the genomic breakpoints occur in intron 9 of BRAF, which generates a truncated protein that dimerize in a RAS-independent manner [148]. Thus, BRAF-KDD adopts a complete different activating mechanism from EGFR-KDD, which give us important clues that possibly KDD in different RTKs use different activation mechanisms. Systematic functional studies of each of the novel identified KDD within RTK are very necessary for the understanding of the entire RTK paradigm.
Autocrine activation
Cell-cell communication utilizes “messengers” – such as growth factors and cytokines – that are released by secretory cells and delivered to remote target cells. “Autocrine” refers to the situation that the target cells are secreting cells themselves [149]. Constitutive autocrine activation might lead to clonal expansion and tumor formation (Fig. 2c) [150], and autocrine activation of various RTKs has been well characterized in diverse cancers, including TGFα-EGFR [151], HGF-MET [152, 153], and SCF-KIT autocrine loops [154,155,156]. RTK autocrine loop may work synergistically with other autocrine growth pathway and drive tumor development. The growth advantage conferred by SCF-KIT loop partially synergizes with another two autocrine loops, IGF-l and bombesin, to drive the development of small cell lung cancer (SCLC) [154]. Autocrine pathways could act as a rational target for cancer therapy [151]. For example, ligand/receptor autocrine loops renders EGFR-mutant lung cancer cells less sensitive to EGFR TKI inhibition [157].
Emerging mechanisms to aberrantly activate RTKs
MicroRNAs
MicroRNAs can directly modulate the expression of RTKs, and function as both tumor suppressors and oncogenes [158]. For example, microRNA-10a promotes metastasis by directly regulating EPH4A-mediated epithelial-mesenchymal transition and adhesion in hepatocellular carcinoma [159]. MicroRNA-145 suppresses the development of lung adenocarcinoma through directly modulating EGFR expressions at both mRNA and protein levels [160]. MicroRNA-219-5p suppresses GBM development through repressing EGFR expression by directly binding to its 3’-UTR [161]. In addition, microRNAs have also been shown to be involved in the RTK signaling and regulation of tumor formation. Recent data has demonstrated that RTKs, such as MET, EGFR, and PDGFR, regulate microRNA-134 in GBM, while microRNA-134 acts as a tumor-suppressive hub and controls KRAS and STAT5B expression levels [162]. Insights into oncogenic microRNAs and RTK signaling will allow exploiting and improving cancer therapies. For example, the combination of a monoclonal antibody against EGFR and an inhibitor of microRNA-21 improve the treatment outcome in GBM [163]. Moreover, microRNAs could function as potential prognostic markers and assist in patient stratification. The microRNA signature (MiR-99a/Let-7c/miR-125b) may serve as biomarker for prognosis of patients with colorectal cancer treated with anti-EGFR antibodies [164]. An improved understanding of microRNAs involved in RTK signaling may have future implications in cancer detection, therapy and prognosis.
Alterations in tumor microenvironment
Several notable advances have been made during the last decade in the recognition of the importance of tumor microenvironment, especially tumor vasculature and tumor stroma [165]. Members of the Eph receptor family mediate cell-cell interaction in tumor stroma and tumor vasculature [166]. Macrophages function as key cellular components of tumor microenvironment. AXL is highly expressed within tumor associated macrophages where AXL may promote immunosuppressive and pre-neoplasia phenotypes [167]. RET and GFRA1 have been shown to be expressed in stromal cells of the bone marrow microenvironment and implicated in the development of acute myeloid leukemias [168]. Many other RTKs have been shown to be important in the tumor microenvironment, including VEGFR [169, 170] and PDGFR [171]. As such, these RTKs represent attractive potential targets for drug design. Many AXL inhibitors have been detected and are efficacious in preclinical studies against cancer [167].
Signal attenuation by negative regulators
The activity of RTKs must be tightly regulated and properly balanced in order to mediate their normal cellular activities and physiological processes. Signal attenuation and downregulation of RTK pathways provide important implications in cancer therapeutics and several well characterized negative regulators in RTK signaling (such as PTEN, LRIG1 and ERRFI1) are bona fide tumor suppressors [172,173,174].
ERRFI1 (ErbB Receptor Feedback Inhibitor 1) – which encodes the protein MIG6 – is located within chromosome 1p36.1–3, a hotspot region frequently deleted in a broad range of human cancers, including breast, liver and kidney cancers [175]. MIG6 has been described to be mutated in different human cancers [176, 177]. MIG6 expression is also downregulated or silenced in skin, breast, pancreatic and ovarian carcinomas [178, 179]. Loss of Errfi1 in mice leads to abnormal activation of EGFR signaling and is associated with a high incidence of neoplastic lesions [178]. These findings suggested that MIG6 played tumor suppressive roles possibly involved in EGFR signaling. MIG6 contains two functional regions, termed segments 1 and 2 which are 77 amino acids in total [174]. Structural studies indicate that MIG6 (segment 1) is able to inhibit EGFR kinase activity in the presence of the asymmetric dimer. MIG6 (segment 1) binds to ‘activator’ kinase and prevents the activation of EGFR, while segment 2 is required for the inhibition of the kinase activity of activated EGFR, and that both segments 1 and 2 are essential for the potent inhibition of EGFR activity [174]. Residues in the binding interface between EGFR and MIG6 (segment 1) are conserved across all ErbB family members rather than other protein kinases [9], However, in another structural study, MIG6 could not effectively inhibit the oncogenic mutants of EGFR (e.g. L858R), presumably because EGFR mutants can form asymmetric dimers at a lower energetic cost than wild-type EGFR [36]. The C-lobe is less accessible by MIG6 in configurations that more strongly favor formation of asymmetric dimers [32]. These two studies give us clues that MIG6 may potentially inhibit EGFR-KDD, EGFR-RAD51 and EGFR-PURB, because these EGFR mutant proteins have intact wild-type TKD which could potentially act as ‘activator’ kinase in the form of activating asymmetric dimerization.
RTKs as therapeutic targets
Since RTKs play crucial roles in cancer development, targeting oncogenic driver mutations of RTKs has revolutionized the treatment of cancer patients. Above, we touched on how targeted therapies are deployed in specific clinical scenarios for patients whose tumors harbor oncogenic RTK variants. However, a detailed review of all RTK inhibitors in the treatment of human tumors is beyond the scope of this manuscript. In brief, many small-molecule inhibitors have been developed for treating cancers and other diseases caused by driver mutations within RTKs. These inhibitors specifically target the ATP-binding site of the intracellular TKD [180]. In addition, the US FDA has approved many monoclonal antibodies that interfere with RTK activation, including cetuximab in lung cancer [181], panitumumab in colon cancer [182], cetuximab in head and neck cancer [183], trastuzumab and pertuzumab in breast cancer [184, 185]. Overall, the development and routine clinical implementation of agents (TKIs and monoclonal antibodies) targeting RTKs has heralded the new age of precision cancer medicine. Despite these advancements, acquired resistance to targeted therapies inevitably develops [40, 133]. Acquired resistance can occur through either acquired genomic alterations [186, 187] or activation of critical signaling pathways [188,189,190]. Novel approaches have been shown to effectively overcome acquired resistance, including the development of second-generation [191, 192] and third-generation inhibitors [193, 194] and the combinational use of TKIs with monoclonal antibodies against the same target [195].
Conclusions
Our understanding of RTK signaling has advanced dramatically in the past two decades. Studies of RTKs have provided fundamental insight into how this protein family functions and how to develop targeted therapeutics. However, much work is still required to fully understand all members of the RTK family. An improved understanding of RTK signaling pathways will provide a strong foundation on which improvements to patient care can be made. An integrated approach, combining genetic, cellular, biochemical, and structural modeling techniques, may offer the most complete view yet of this critical family of protein tyrosine kinases.
Abbreviations
- ALCL:
-
Anaplastic large cell lymphoma;
- CML:
-
Chronic myelogenous leukemia
- ECD:
-
Extracellular domain
- FDA:
-
Food and Drug Administration
- GBM:
-
Glioblastoma
- GIST:
-
Gastrointestinal stromal tumor
- IMT:
-
Inflammatory myofibroblastic tumor
- IR:
-
Insulin receptor
- JMD:
-
Juxtamembrane domain
- KDD:
-
Kinase domain duplication
- NGS:
-
Next generation sequencing
- NSCLC:
-
Non-small cell lung cancer
- PTB:
-
Phosphotyrosine-binding domain
- RTK:
-
Receptor tyrosine kinases
- SCLC:
-
Small cell lung cancer
- SH2:
-
Src homology-2 domain
- TKD:
-
Tyrosine kinase domain
- TKI:
-
Tyrosine kinase inhibitor
- TMD:
-
Transmembrane domain
References
Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–34.
Robinson DR, Wu YM, Lin SF. The protein tyrosine kinase family of the human genome. Oncogene. 2000;19:5548–57.
Hubbard SR. Structural analysis of receptor tyrosine kinases. Prog Biophys Mol Biol. 1999;71:343–58.
Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–58.
Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–25.
Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–34.
Pawson T, Gish GD, Nash P. SH2 domains, interaction modules and cellular wiring. Trends Cell Biol. 2001;11:504–11.
Wehrman T, He X, Raab B, Dukipatti A, Blau H, Garcia KC. Structural and mechanistic insights into nerve growth factor interactions with the TrkA and p75 receptors. Neuron. 2007;53:25–38.
Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–49.
Yuzawa S, Opatowsky Y, Zhang Z, Mandiyan V, Lax I, Schlessinger J. Structural basis for activation of the receptor tyrosine kinase KIT by stem cell factor. Cell. 2007;130:323–34.
Yayon A, Klagsbrun M, Esko JD, Leder P, Ornitz DM. Cell surface, heparin-like molecules are required for binding of basic fibroblast growth factor to its high affinity receptor. Cell. 1991;64:841–8.
Schlessinger J, Plotnikov AN, Ibrahimi OA, Eliseenkova AV, Yeh BK, Yayon A, et al. Crystal structure of a ternary FGF-FGFR-heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol Cell. 2000;6:743–50.
Chung I, Akita R, Vandlen R, Toomre D, Schlessinger J, Mellman I. Spatial control of EGF receptor activation by reversible dimerization on living cells. Nature. 2010;464:783–7.
Soos MA, Field CE, Siddle K. Purified hybrid insulin/insulin-like growth factor-I receptors bind insulin-like growth factor-I, but not insulin, with high affinity. Biochem J. 1993;290(Pt 2):419–26.
Pandini G, Frasca F, Mineo R, Sciacca L, Vigneri R, Belfiore A. Insulin/insulin-like growth factor I hybrid receptors have different biological characteristics depending on the insulin receptor isoform involved. J Biol Chem. 2002;277:39684–95.
Ogiso H, Ishitani R, Nureki O, Fukai S, Yamanaka M, Kim JH, et al. Crystal structure of the complex of human epidermal growth factor and receptor extracellular domains. Cell. 2002;110:775–87.
Stauber DJ, DiGabriele AD, Hendrickson WA. Structural interactions of fibroblast growth factor receptor with its ligands. Proc Natl Acad Sci U S A. 2000;97:49–54.
Burgess AW, Cho HS, Eigenbrot C, Ferguson KM, Garrett TP, Leahy DJ, et al. An open-and-shut case? Recent insights into the activation of EGF/ErbB receptors. Mol Cell. 2003;12:541–52.
Shewchuk LM, Hassell AM, Ellis B, Holmes WD, Davis R, Horne EL, et al. Structure of the Tie2 RTK domain: self-inhibition by the nucleotide binding loop, activation loop, and C-terminal tail. Structure. 2000;8:1105–13.
Wybenga-Groot LE, Baskin B, Ong SH, Tong J, Pawson T, Sicheri F. Structural basis for autoinhibition of the Ephb2 receptor tyrosine kinase by the unphosphorylated juxtamembrane region. Cell. 2001;106:745–57.
Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–82.
Nolen B, Taylor S, Ghosh G. Regulation of protein kinases; controlling activity through activation segment conformation. Mol Cell. 2004;15:661–75.
Mol CD, Dougan DR, Schneider TR, Skene RJ, Kraus ML, Scheibe DN, et al. Structural basis for the autoinhibition and STI-571 inhibition of c-kit tyrosine kinase. J Biol Chem. 2004;279:31655–63.
Red Brewer M, Choi SH, Alvarado D, Moravcevic K, Pozzi A, Lemmon MA, et al. The juxtamembrane region of the EGF receptor functions as an activation domain. Mol Cell. 2009;34:641–51.
Jura N, Endres NF, Engel K, Deindl S, Das R, Lamers MH, et al. Mechanism for activation of the EGF receptor catalytic domain by the juxtamembrane segment. Cell. 2009;137:1293–307.
Brummer T, Schmitz-Peiffer C, Daly RJ. Docking proteins. FEBS J. 2010;277:4356–69.
Ostman A, Bohmer FD. Regulation of receptor tyrosine kinase signaling by protein tyrosine phosphatases. Trends Cell Biol. 2001;11:258–66.
Casaletto JB, McClatchey AI. Spatial regulation of receptor tyrosine kinases in development and cancer. Nat Rev Cancer. 2012;12:387–400.
McDonell LM, Kernohan KD, Boycott KM, Sawyer SL. Receptor tyrosine kinase mutations in developmental syndromes and cancer: two sides of the same coin. Hum Mol Genet. 2015;24:R60–6.
Lahiry P, Torkamani A, Schork NJ, Hegele RA. Kinase mutations in human disease: interpreting genotype-phenotype relationships. Nat Rev Genet. 2010;11:60–74.
Medves S, Demoulin JB. Tyrosine kinase gene fusions in cancer: translating mechanisms into targeted therapies. J Cell Mol Med. 2012;16:237–48.
Wang Z, Longo PA, Tarrant MK, Kim K, Head S, Leahy DJ, et al. Mechanistic insights into the activation of oncogenic forms of EGF receptor. Nat Struct Mol Biol. 2011;18:1388–93.
Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169–81.
Janne PA, Engelman JA, Johnson BE. Epidermal growth factor receptor mutations in non-small-cell lung cancer: implications for treatment and tumor biology. J Clin Oncol. 2005;23:3227–34.
Marchetti A, Martella C, Felicioni L, Barassi F, Salvatore S, Chella A, et al. EGFR mutations in non-small-cell lung cancer: analysis of a large series of cases and development of a rapid and sensitive method for diagnostic screening with potential implications on pharmacologic treatment. J Clin Oncol. 2005;23:857–65.
Red Brewer M, Yun CH, Lai D, Lemmon MA, Eck MJ, Pao W. Mechanism for activation of mutated epidermal growth factor receptors in lung cancer. Proc Natl Acad Sci U S A. 2013;110:E3595–604.
Yun CH, Boggon TJ, Li Y, Woo MS, Greulich H, Meyerson M, et al. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell. 2007;11:217–27.
Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13:239–46.
Zhou C, Wu YL, Chen G, Feng J, Liu XQ, Wang C, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12:735–42.
Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–57.
Mitsudomi T, Morita S, Yatabe Y, Negoro S, Okamoto I, Tsurutani J, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol. 2010;11:121–8.
Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–8.
Sequist LV, Yang JC, Yamamoto N, O'Byrne K, Hirsh V, Mok T, et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol. 2013;31:3327–34.
Janne PA, Yang JC, Kim DW, Planchard D, Ohe Y, Ramalingam SS, et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med. 2015;372:1689–99.
Soria JC, Ohe Y, Vansteenkiste J, Reungwetwattana T, Chewaskulyong B, Lee KH, et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N Engl J Med. 2018;378:113–25.
Lee JC, Vivanco I, Beroukhim R, Huang JH, Feng WL, DeBiasi RM, et al. Epidermal growth factor receptor activation in glioblastoma through novel missense mutations in the extracellular domain. PLoS Med. 2006;3:e485.
Arjona D, Bello MJ, Alonso ME, Aminoso C, Isla A, De Campos JM, et al. Molecular analysis of the EGFR gene in astrocytic gliomas: mRNA expression, quantitative-PCR analysis of non-homogeneous gene amplification and DNA sequence alterations. Neuropathol Appl Neurobiol. 2005;31:384–94.
van den Bent MJ, Brandes AA, Rampling R, Kouwenhoven MC, Kros JM, Carpentier AF, et al. Randomized phase II trial of erlotinib versus temozolomide or carmustine in recurrent glioblastoma: EORTC brain tumor group study 26034. J Clin Oncol. 2009;27:1268–74.
Franceschi E, Cavallo G, Lonardi S, Magrini E, Tosoni A, Grosso D, et al. Gefitinib in patients with progressive high-grade gliomas: a multicentre phase II study by Gruppo Italiano Cooperativo di neuro-Oncologia (GICNO). Br J Cancer. 2007;96:1047–51.
Vivanco I, Robins HI, Rohle D, Campos C, Grommes C, Nghiemphu PL, et al. Differential sensitivity of glioma- versus lung cancer-specific EGFR mutations to EGFR kinase inhibitors. Cancer Discov. 2012;2:458–71.
Wu R, Connolly D, Ngelangel C, Bosch FX, Munoz N, Cho KR. Somatic mutations of fibroblast growth factor receptor 3 (FGFR3) are uncommon in carcinomas of the uterine cervix. Oncogene. 2000;19:5543–6.
Robertson SC, Meyer AN, Hart KC, Galvin BD, Webster MK, Donoghue DJ. Activating mutations in the extracellular domain of the fibroblast growth factor receptor 2 function by disruption of the disulfide bond in the third immunoglobulin-like domain. Proc Natl Acad Sci U S A. 1998;95:4567–72.
Tallini G, Asa SL. RET oncogene activation in papillary thyroid carcinoma. Adv Anat Pathol. 2001;8:345–54.
Heinrich MC, Corless CL, Demetri GD, Blanke CD, von Mehren M, Joensuu H, et al. Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J Clin Oncol. 2003;21:4342–9.
Ou SI, Schrock AB, Bocharov EV, Klempner SJ, Haddad CK, Steinecker G, et al. HER2 transmembrane domain (TMD) mutations (V659/G660) that stabilize homo- and Heterodimerization are rare oncogenic drivers in lung adenocarcinoma that respond to Afatinib. J Thorac Oncol. 2017;12:446–57.
Serra V, Vivancos A, Puente XS, Felip E, Silberschmidt D, Caratu G, et al. Clinical response to a lapatinib-based therapy for a li-Fraumeni syndrome patient with a novel HER2V659E mutation. Cancer Discov. 2013;3:1238–44.
Bocharov EV, Lesovoy DM, Pavlov KV, Pustovalova YE, Bocharova OV, Arseniev AS. Alternative packing of EGFR transmembrane domain suggests that protein-lipid interactions underlie signal conduction across membrane. Biochim Biophys Acta. 1858;2016:1254–61.
Yamamoto H, Higasa K, Sakaguchi M, Shien K, Soh J, Ichimura K, et al. Novel germline mutation in the transmembrane domain of HER2 in familial lung adenocarcinomas. J Natl Cancer Inst. 2014;106:djt338.
Yamamoto H, Toyooka S, Ninomiya T, Matsumoto S, Kanai M, Tomida S, et al. Therapeutic Potential of Afatinib for Cancers with ERBB2 (HER2) Transmembrane Domain Mutations G660D and V659E. Oncologist. 2017. https://doi.org/10.1634/theoncologist.2017-0345.
Heinrich MC, Griffith DJ, Druker BJ, Wait CL, Ott KA, Zigler AJ. Inhibition of c-kit receptor tyrosine kinase activity by STI 571, a selective tyrosine kinase inhibitor. Blood. 2000;96:925–32.
Buchdunger E, Cioffi CL, Law N, Stover D, Ohno-Jones S, Druker BJ, et al. Abl protein-tyrosine kinase inhibitor STI571 inhibits in vitro signal transduction mediated by c-kit and platelet-derived growth factor receptors. J Pharmacol Exp Ther. 2000;295:139–45.
Lopez-Gines C, Gil-Benso R, Ferrer-Luna R, Benito R, Serna E, Gonzalez-Darder J, et al. New pattern of EGFR amplification in glioblastoma and the relationship of gene copy number with gene expression profile. Mod Pathol. 2010;23:856–65.
Selvaggi G, Novello S, Torri V, Leonardo E, De Giuli P, Borasio P, et al. Epidermal growth factor receptor overexpression correlates with a poor prognosis in completely resected non-small-cell lung cancer. Ann Oncol. 2004;15:28–32.
Hanawa M, Suzuki S, Dobashi Y, Yamane T, Kono K, Enomoto N, et al. EGFR protein overexpression and gene amplification in squamous cell carcinomas of the esophagus. Int J Cancer. 2006;118:1173–80.
Rodriguez-Antona C, Pallares J, Montero-Conde C, Inglada-Perez L, Castelblanco E, Landa I, et al. Overexpression and activation of EGFR and VEGFR2 in medullary thyroid carcinomas is related to metastasis. Endocr Relat Cancer. 2010;17:7–16.
Hirsch FR, Varella-Garcia M, Cappuzzo F. Predictive value of EGFR and HER2 overexpression in advanced non-small-cell lung cancer. Oncogene. 2009;28(Suppl 1):S32–7.
Menard S, Casalini P, Campiglio M, Pupa S, Agresti R, Tagliabue E. HER2 overexpression in various tumor types, focussing on its relationship to the development of invasive breast cancer. Ann Oncol. 2001;12(Suppl 1):S15–9.
Yaziji H, Goldstein LC, Barry TS, Werling R, Hwang H, Ellis GK, et al. HER-2 testing in breast cancer using parallel tissue-based methods. JAMA. 2004;291:1972–7.
Kim KC, Koh YW, Chang HM, Kim TH, Yook JH, Kim BS, et al. Evaluation of HER2 protein expression in gastric carcinomas: comparative analysis of 1,414 cases of whole-tissue sections and 595 cases of tissue microarrays. Ann Surg Oncol. 2011;18:2833–40.
Park DI, Yun JW, Park JH, Oh SJ, Kim HJ, Cho YK, et al. HER-2/neu amplification is an independent prognostic factor in gastric cancer. Dig Dis Sci. 2006;51:1371–9.
Xu L, Nilsson MB, Saintigny P, Cascone T, Herynk MH, Du Z, et al. Epidermal growth factor receptor regulates MET levels and invasiveness through hypoxia-inducible factor-1alpha in non-small cell lung cancer cells. Oncogene. 2010;29:2616–27.
Ha SY, Lee J, Kang SY, Do IG, Ahn S, Park JO, et al. MET overexpression assessed by new interpretation method predicts gene amplification and poor survival in advanced gastric carcinomas. Mod Pathol. 2013;26:1632–41.
Carraway KL 3rd, Sweeney C. EGF receptor activation by heterologous mechanisms. Cancer Cell. 2002;1:405–6.
Ludes-Meyers JH, Subler MA, Shivakumar CV, Munoz RM, Jiang P, Bigger JE, et al. Transcriptional activation of the human epidermal growth factor receptor promoter by human p53. Mol Cell Biol. 1996;16:6009–19.
Reznik TE, Sang Y, Ma Y, Abounader R, Rosen EM, Xia S, et al. Transcription-dependent epidermal growth factor receptor activation by hepatocyte growth factor. Mol Cancer Res. 2008;6:139–50.
Sun T, Aceto N, Meerbrey KL, Kessler JD, Zhou C, Migliaccio I, et al. Activation of multiple proto-oncogenic tyrosine kinases in breast cancer via loss of the PTPN12 phosphatase. Cell. 2011;144:703–18.
Maiti GP, Mondal P, Mukherjee N, Ghosh A, Ghosh S, Dey S, et al. Overexpression of EGFR in head and neck squamous cell carcinoma is associated with inactivation of SH3GL2 and CDC25A genes. PLoS One. 2013;8:e63440.
Mudduluru G, Ceppi P, Kumarswamy R, Scagliotti GV, Papotti M, Allgayer H. Regulation of Axl receptor tyrosine kinase expression by miR-34a and miR-199a/b in solid cancer. Oncogene. 2011;30:2888–99.
Templeton AJ, Diez-Gonzalez L, Ace O, Vera-Badillo F, Seruga B, Jordan J, et al. Prognostic relevance of receptor tyrosine kinase expression in breast cancer: a meta-analysis. Cancer Treat Rev. 2014;40:1048–55.
Albertson DG. Gene amplification in cancer. Trends Genet. 2006;22:447–55.
Albertson DG, Collins C, McCormick F, Gray JW. Chromosome aberrations in solid tumors. Nat Genet. 2003;34:369–76.
Dutt A, Ramos AH, Hammerman PS, Mermel C, Cho J, Sharifnia T, et al. Inhibitor-sensitive FGFR1 amplification in human non-small cell lung cancer. PLoS One. 2011;6:e20351.
Reis-Filho JS, Simpson PT, Turner NC, Lambros MB, Jones C, Mackay A, et al. FGFR1 emerges as a potential therapeutic target for lobular breast carcinomas. Clin Cancer Res. 2006;12:6652–62.
Helsten T, Elkin S, Arthur E, Tomson BN, Carter J, Kurzrock R. The FGFR landscape in cancer: analysis of 4,853 tumors by next-generation sequencing. Clin Cancer Res. 2016;22:259–67.
Fischbach A, Rogler A, Erber R, Stoehr R, Poulsom R, Heidenreich A, et al. Fibroblast growth factor receptor (FGFR) gene amplifications are rare events in bladder cancer. Histopathology. 2015;66:639–49.
Kim JY, Jung HH, Do IG, Bae S, Lee SK, Kim SW, et al. Prognostic value of ERBB4 expression in patients with triple negative breast cancer. BMC Cancer. 2016;16:138.
Shi J, Yao D, Liu W, Wang N, Lv H, He N, et al. Frequent gene amplification predicts poor prognosis in gastric cancer. Int J Mol Sci. 2012;13:4714–26.
Moreira RB, Peixoto RD, de Sousa Cruz MR. Clinical response to Sorafenib in a patient with metastatic colorectal cancer and FLT3 amplification. Case Rep Oncol. 2015;8:83–7.
Carvajal RD, Antonescu CR, Wolchok JD, Chapman PB, Roman RA, Teitcher J, et al. KIT as a therapeutic target in metastatic melanoma. JAMA. 2011;305:2327–34.
Tabone S, Theou N, Wozniak A, Saffroy R, Deville L, Julie C, et al. KIT overexpression and amplification in gastrointestinal stromal tumors (GISTs). Biochim Biophys Acta. 2005;1741:165–72.
Nobusawa S, Stawski R, Kim YH, Nakazato Y, Ohgaki H. Amplification of the PDGFRA, KIT and KDR genes in glioblastoma: a population-based study. Neuropathology. 2011;31:583–8.
Sholl LM, Yeap BY, Iafrate AJ, Holmes-Tisch AJ, Chou YP, Wu MT, et al. Lung adenocarcinoma with EGFR amplification has distinct clinicopathologic and molecular features in never-smokers. Cancer Res. 2009;69:8341–8.
Yu HA, Arcila ME, Rekhtman N, Sima CS, Zakowski MF, Pao W, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res. 2013;19:2240–7.
Stransky N, Cerami E, Schalm S, Kim JL, Lengauer C. The landscape of kinase fusions in cancer. Nat Commun. 2014;5:4846.
Cancer Genome Atlas Research N. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–50.
Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–77.
Nowell PC. Discovery of the Philadelphia chromosome: a personal perspective. J Clin Invest. 2007;117:2033–5.
Diamond J, Goldman JM, Melo JV. BCR-ABL, ABL-BCR, BCR, and ABL genes are all expressed in individual granulocyte-macrophage colony-forming unit colonies derived from blood of patients with chronic myeloid leukemia. Blood. 1995;85:2171–5.
Melo JV, Gordon DE, Cross NC, Goldman JM. The ABL-BCR fusion gene is expressed in chronic myeloid leukemia. Blood. 1993;81:158–65.
Druker BJ, Talpaz M, Resta DJ, Peng B, Buchdunger E, Ford JM, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344:1031–7.
O'Brien SG, Guilhot F, Larson RA, Gathmann I, Baccarani M, Cervantes F, et al. Imatinib compared with interferon and low-dose cytarabine for newly diagnosed chronic-phase chronic myeloid leukemia. N Engl J Med. 2003;348:994–1004.
Morris SW, Kirstein MN, Valentine MB, Dittmer KG, Shapiro DN, Saltman DL, et al. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin's lymphoma. Science. 1994;263:1281–4.
Kutok JL, Aster JC. Molecular biology of anaplastic lymphoma kinase-positive anaplastic large-cell lymphoma. J Clin Oncol. 2002;20:3691–702.
Takeuchi K, Soda M, Togashi Y, Suzuki R, Sakata S, Hatano S, et al. RET, ROS1 and ALK fusions in lung cancer. Nat Med. 2012;18:378–81.
Coffin CM, Hornick JL, Fletcher CD. Inflammatory myofibroblastic tumor: comparison of clinicopathologic, histologic, and immunohistochemical features including ALK expression in atypical and aggressive cases. Am J Surg Pathol. 2007;31:509–20.
Lovly CM, Gupta A, Lipson D, Otto G, Brennan T, Chung CT, et al. Inflammatory myofibroblastic tumors harbor multiple potentially actionable kinase fusions. Cancer Discov. 2014;4:889–95.
Wiesner T, He J, Yelensky R, Esteve-Puig R, Botton T, Yeh I, et al. Kinase fusions are frequent in Spitz tumours and spitzoid melanomas. Nat Commun. 2014;5:3116.
Lipson D, Capelletti M, Yelensky R, Otto G, Parker A, Jarosz M, et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med. 2012;18:382–4.
Lai AZ, Schrock AB, Erlich RL, Ross JS, Miller VA, Yakirevich E, et al. Detection of an ALK fusion in colorectal carcinoma by hybrid capture-based assay of circulating tumor DNA. Oncologist. 2017;22:774–9.
Chou A, Fraser S, Toon CW, Clarkson A, Sioson L, Farzin M, et al. A detailed clinicopathologic study of ALK-translocated papillary thyroid carcinoma. Am J Surg Pathol. 2015;39:652–9.
Ren H, Tan ZP, Zhu X, Crosby K, Haack H, Ren JM, et al. Identification of anaplastic lymphoma kinase as a potential therapeutic target in ovarian cancer. Cancer Res. 2012;72:3312–23.
Bergethon K, Shaw AT, Ou SH, Katayama R, Lovly CM, McDonald NT, et al. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol. 2012;30:863–70.
Uguen A, De Braekeleer M. ROS1 fusions in cancer: a review. Future Oncol. 2016;12:1911–28.
Dacic S, Luvison A, Evdokimova V, Kelly L, Siegfried JM, Villaruz LC, et al. RET rearrangements in lung adenocarcinoma and radiation. J Thorac Oncol. 2014;9:118–20.
Nikiforov YE. RET/PTC rearrangement in thyroid tumors. Endocr Pathol. 2002;13:3–16.
Hyman DM, Laetsch TW, Kummar S, DuBois SG, Farago AF, Pappo AS, et al. The efficacy of larotrectinib (LOXO-101), a selective tropomyosin receptor kinase (TRK) inhibitor, in adult and pediatric TRK fusion cancers. J Clin Oncol. 2017. https://doi.org/10.1200/JCO.2017.35.15_suppl.LBA2501.
Konduri K, Gallant JN, Chae YK, Giles FJ, Gitlitz BJ, Gowen K, et al. EGFR fusions as novel therapeutic targets in lung cancer. Cancer Discov. 2016;6:601–11.
Chmielecki J, Ross JS, Wang K, Frampton GM, Palmer GA, Ali SM, et al. Oncogenic alterations in ERBB2/HER2 represent potential therapeutic targets across tumors from diverse anatomic sites of origin. Oncologist. 2015;20:7–12.
Huang Q, Snyder DS, Chu P, Gaal KK, Chang KL, Weiss LM. PDGFRA rearrangement leading to hyper-eosinophilia, T-lymphoblastic lymphoma, myeloproliferative neoplasm and precursor B-cell acute lymphoblastic leukemia. Leukemia. 2011;25:371–5.
Ito T, Seyama T, Iwamoto KS, Hayashi T, Mizuno T, Tsuyama N, et al. In vitro irradiation is able to cause RET oncogene rearrangement. Cancer Res. 1993;53:2940–3.
Mizuno T, Kyoizumi S, Suzuki T, Iwamoto KS, Seyama T. Continued expression of a tissue specific activated oncogene in the early steps of radiation-induced human thyroid carcinogenesis. Oncogene. 1997;15:1455–60.
Mistry AR, Felix CA, Whitmarsh RJ, Mason A, Reiter A, Cassinat B, et al. DNA topoisomerase II in therapy-related acute promyelocytic leukemia. N Engl J Med. 2005;352:1529–38.
Tsai AG, Lieber MR. Mechanisms of chromosomal rearrangement in the human genome. BMC Genomics. 2010;11(Suppl 1):S1.
Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–6.
Noh KW, Lee MS, Lee SE, Song JY, Shin HT, Kim YJ, et al. Molecular breakdown: a comprehensive view of anaplastic lymphoma kinase (ALK)-rearranged non-small cell lung cancer. J Pathol. 2017;243:307–19.
Ju YS, Lee WC, Shin JY, Lee S, Bleazard T, Won JK, et al. A transforming KIF5B and RET gene fusion in lung adenocarcinoma revealed from whole-genome and transcriptome sequencing. Genome Res. 2012;22:436–45.
Wu YM, Su F, Kalyana-Sundaram S, Khazanov N, Ateeq B, Cao X, et al. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov. 2013;3:636–47.
Ross TS, Gilliland DG. Transforming properties of the huntingtin interacting protein 1/ platelet-derived growth factor beta receptor fusion protein. J Biol Chem. 1999;274:22328–36.
Martelli MP, Sozzi G, Hernandez L, Pettirossi V, Navarro A, Conte D, et al. EML4-ALK rearrangement in non-small cell lung cancer and non-tumor lung tissues. Am J Pathol. 2009;174:661–70.
Corvi R, Berger N, Balczon R, Romeo G. RET/PCM-1: a novel fusion gene in papillary thyroid carcinoma. Oncogene. 2000;19:4236–42.
Iragavarapu C, Mustafa M, Akinleye A, Furqan M, Mittal V, Cang S, et al. Novel ALK inhibitors in clinical use and development. J Hematol Oncol. 2015;8:17.
Butrynski JE, D'Adamo DR, Hornick JL, Dal Cin P, Antonescu CR, Jhanwar SC, et al. Crizotinib in ALK-rearranged inflammatory myofibroblastic tumor. N Engl J Med. 2010;363:1727–33.
Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–703.
Kloos RT, Ringel MD, Knopp MV, Hall NC, King M, Stevens R, et al. Phase II trial of sorafenib in metastatic thyroid cancer. J Clin Oncol. 2009;27:1675–84.
Wang R, Hu H, Pan Y, Li Y, Ye T, Li C, et al. RET fusions define a unique molecular and clinicopathologic subtype of non-small-cell lung cancer. J Clin Oncol. 2012;30:4352–9.
Drilon A, Wang L, Hasanovic A, Suehara Y, Lipson D, Stephens P, et al. Response to Cabozantinib in patients with RET fusion-positive lung adenocarcinomas. Cancer Discov. 2013;3:630–5.
Drilon A, Rekhtman N, Arcila M, Wang L, Ni A, Albano M, et al. Cabozantinib in patients with advanced RET-rearranged non-small-cell lung cancer: an open-label, single-centre, phase 2, single-arm trial. Lancet Oncol. 2016;17:1653–60.
Kiehna EN, Arnush MR, Tamrazi B, Cotter JA, Hawes D, Robison NJ, et al. Novel GOPC(FIG)-ROS1 fusion in a pediatric high-grade glioma survivor. J Neurosurg Pediatr. 2017;20:51–5.
Shaw AT, Solomon BJ. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med. 2015;372:683–4.
Drilon A, Nagasubramanian R, Blake JF, Ku N, Tuch BB, Ebata K, et al. A next-generation TRK kinase inhibitor overcomes acquired resistance to prior TRK kinase inhibition in patients with TRK fusion-positive solid tumors. Cancer Discov. 2017;7:963–72.
Drilon A, Siena S, Ou SI, Patel M, Ahn MJ, Lee J, et al. Safety and antitumor activity of the multitargeted pan-TRK, ROS1, and ALK inhibitor Entrectinib: combined results from two phase I trials (ALKA-372-001 and STARTRK-1). Cancer Discov. 2017;7:400–9.
Chen HY, Brady DC, Villanueva J. Double trouble: kinase domain duplication as a new path to drug resistance. Pigment Cell Melanoma Res. 2016;29:493–5.
Gallant JN, Sheehan JH, Shaver TM, Bailey M, Lipson D, Chandramohan R, et al. EGFR kinase domain duplication (EGFR-KDD) is a novel oncogenic driver in lung cancer that is clinically responsive to Afatinib. Cancer Discov. 2015;5:1155–63.
Rodriguez FJ, Ligon AH, Horkayne-Szakaly I, Rushing EJ, Ligon KL, Vena N, et al. BRAF duplications and MAPK pathway activation are frequent in gliomas of the optic nerve proper. J Neuropathol Exp Neurol. 2012;71:789–94.
Klempner SJ, Bordoni R, Gowen K, Kaplan H, Stephens PJ, Ou SH, et al. Identification of BRAF kinase domain duplications across multiple tumor types and response to RAF inhibitor therapy. JAMA Oncol. 2016;2:272–4.
Gay LM, Pavlick D, Chung J, Ramkissoon S, Daniel S, Elvin JA, et al. Genomic profiling of 114,200 advanced cancers identifies recurrent kinase domain duplications (KDD) and oncogenic rearrangements (RE) across diverse tumor types. Ann Oncol. 2017;28:v595–604.
Kondrashov FA. Gene duplication as a mechanism of genomic adaptation to a changing environment. Proc Biol Sci. 2012;279:5048–57.
Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E). Nature. 2011;480:387–90.
Singh AB, Harris RC. Autocrine, paracrine and juxtacrine signaling by EGFR ligands. Cell Signal. 2005;17:1183–93.
Walsh JH, Karnes WE, Cuttitta F, Walker A. Autocrine growth factors and solid tumor malignancy. West J Med. 1991;155:152–63.
Ciardiello F, Tortora G. A novel approach in the treatment of cancer: targeting the epidermal growth factor receptor. Clin Cancer Res. 2001;7:2958–70.
Kentsis A, Reed C, Rice KL, Sanda T, Rodig SJ, Tholouli E, et al. Autocrine activation of the MET receptor tyrosine kinase in acute myeloid leukemia. Nat Med. 2012;18:1118–22.
Yi S, Tsao MS. Activation of hepatocyte growth factor-met autocrine loop enhances tumorigenicity in a human lung adenocarcinoma cell line. Neoplasia. 2000;2:226–34.
Krystal GW, Hines SJ, Organ CP. Autocrine growth of small cell lung cancer mediated by coexpression of c-kit and stem cell factor. Cancer Res. 1996;56:370–6.
Wiesner C, Nabha SM, Dos Santos EB, Yamamoto H, Meng H, Melchior SW, et al. C-kit and its ligand stem cell factor: potential contribution to prostate cancer bone metastasis. Neoplasia. 2008;10:996–1003.
Esposito I, Kleeff J, Bischoff SC, Fischer L, Collecchi P, Iorio M, et al. The stem cell factor-c-kit system and mast cells in human pancreatic cancer. Lab Investig. 2002;82:1481–92.
Fujimoto N, Wislez M, Zhang J, Iwanaga K, Dackor J, Hanna AE, et al. High expression of ErbB family members and their ligands in lung adenocarcinomas that are sensitive to inhibition of epidermal growth factor receptor. Cancer Res. 2005;65:11478–85.
Donzelli S, Cioce M, Muti P, Strano S, Yarden Y, Blandino G. MicroRNAs: non-coding fine tuners of receptor tyrosine kinase signalling in cancer. Semin Cell Dev Biol. 2016;50:133–42.
Yan Y, Luo YC, Wan HY, Wang J, Zhang PP, Liu M, et al. MicroRNA-10a is involved in the metastatic process by regulating Eph tyrosine kinase receptor A4-mediated epithelial-mesenchymal transition and adhesion in hepatoma cells. Hepatology. 2013;57:667–77.
Cho WC, Chow AS, Au JS. MiR-145 inhibits cell proliferation of human lung adenocarcinoma by targeting EGFR and NUDT1. RNA Biol. 2011;8:125–31.
Rao SA, Arimappamagan A, Pandey P, Santosh V, Hegde AS, Chandramouli BA, et al. miR-219-5p inhibits receptor tyrosine kinase pathway by targeting EGFR in glioblastoma. PLoS One. 2013;8:e63164.
Zhang Y, Kim J, Mueller AC, Dey B, Yang Y, Lee DH, et al. Multiple receptor tyrosine kinases converge on microRNA-134 to control KRAS, STAT5B, and glioblastoma. Cell Death Differ. 2014;21:720–34.
Zhang KL, Han L, Chen LY, Shi ZD, Yang M, Ren Y, et al. Blockage of a miR-21/EGFR regulatory feedback loop augments anti-EGFR therapy in glioblastomas. Cancer Lett. 2014;342:139–49.
Cappuzzo F, Sacconi A, Landi L, Ludovini V, Biagioni F, D'Incecco A, et al. MicroRNA signature in metastatic colorectal cancer patients treated with anti-EGFR monoclonal antibodies. Clin Colorectal Cancer. 2014;13:37–45. e4
Spill F, Reynolds DS, Kamm RD, Zaman MH. Impact of the physical microenvironment on tumor progression and metastasis. Curr Opin Biotechnol. 2016;40:41–8.
Brantley-Sieders D, Schmidt S, Parker M, Chen J. Eph receptor tyrosine kinases in tumor and tumor microenvironment. Curr Pharm Des. 2004;10:3431–42.
Rankin EB, Giaccia AJ. The Receptor Tyrosine Kinase AXL in Cancer Progression. Cancers (Basel). 2016. https://doi.org/10.3390/cancers8110103.
Gattei V, Celetti A, Cerrato A, Degan M, De Iuliis A, Rossi FM, et al. Expression of the RET receptor tyrosine kinase and GDNFR-alpha in normal and leukemic human hematopoietic cells and stromal cells of the bone marrow microenvironment. Blood. 1997;89:2925–37.
Ribatti D, Ranieri G, Basile A, Azzariti A, Paradiso A, Vacca A. Tumor endothelial markers as a target in cancer. Expert Opin Ther Targets. 2012;16:1215–25.
Bertolini F, Mancuso P, Benayoun L, Gingis-Velitski S, Shaked Y. Evaluation of circulating endothelial precursor cells in cancer patients. Methods Mol Biol. 2012;904:165–72.
Gialeli C, Nikitovic D, Kletsas D, Theocharis AD, Tzanakakis GN, Karamanos NK. PDGF/PDGFR signaling and targeting in cancer growth and progression: focus on tumor microenvironment and cancer-associated fibroblasts. Curr Pharm Des. 2014;20:2843–8.
Worby CA, Dixon JE. PTEN. Annu Rev Biochem. 2014;83:641–69.
Gur G, Rubin C, Katz M, Amit I, Citri A, Nilsson J, et al. LRIG1 restricts growth factor signaling by enhancing receptor ubiquitylation and degradation. EMBO J. 2004;23:3270–81.
Zhang X, Pickin KA, Bose R, Jura N, Cole PA, Kuriyan J. Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface. Nature. 2007;450:741–4.
Bagchi A, Mills AA. The quest for the 1p36 tumor suppressor. Cancer Res. 2008;68:2551–6.
Koshikawa K, Nomoto S, Yamashita K, Ishigure K, Takeda S, Nakao A. Allelic imbalance at 1p36 in the pathogenesis of human hepatocellular carcinoma. Hepato-Gastroenterology. 2004;51:186–91.
Tseng RC, Chang JW, Hsien FJ, Chang YH, Hsiao CF, Chen JT, et al. Genomewide loss of heterozygosity and its clinical associations in non small cell lung cancer. Int J Cancer. 2005;117:241–7.
Ferby I, Reschke M, Kudlacek O, Knyazev P, Pante G, Amann K, et al. Mig6 is a negative regulator of EGF receptor-mediated skin morphogenesis and tumor formation. Nat Med. 2006;12:568–73.
Amatschek S, Koenig U, Auer H, Steinlein P, Pacher M, Gruenfelder A, et al. Tissue-wide expression profiling using cDNA subtraction and microarrays to identify tumor-specific genes. Cancer Res. 2004;64:844–56.
Shawver LK, Slamon D, Ullrich A. Smart drugs: tyrosine kinase inhibitors in cancer therapy. Cancer Cell. 2002;1:117–23.
Pirker R, Pereira JR, Szczesna A, von Pawel J, Krzakowski M, Ramlau R, et al. Cetuximab plus chemotherapy in patients with advanced non-small-cell lung cancer (FLEX): an open-label randomised phase III trial. Lancet. 2009;373:1525–31.
Gibson TB, Ranganathan A, Grothey A. Randomized phase III trial results of panitumumab, a fully human anti-epidermal growth factor receptor monoclonal antibody, in metastatic colorectal cancer. Clin Colorectal Cancer. 2006;6:29–31.
Vermorken JB, Mesia R, Rivera F, Remenar E, Kawecki A, Rottey S, et al. Platinum-based chemotherapy plus cetuximab in head and neck cancer. N Engl J Med. 2008;359:1116–27.
Romond EH, Perez EA, Bryant J, Suman VJ, Geyer CE Jr, Davidson NE, et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med. 2005;353:1673–84.
von Minckwitz G, Procter M, de Azambuja E, Zardavas D, Benyunes M, Viale G, et al. Adjuvant Pertuzumab and Trastuzumab in early HER2-positive breast cancer. N Engl J Med. 2017;377:122–31.
Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–92.
Sasaki T, Koivunen J, Ogino A, Yanagita M, Nikiforow S, Zheng W, et al. A novel ALK secondary mutation and EGFR signaling cause resistance to ALK kinase inhibitors. Cancer Res. 2011;71:6051–60.
Engelman JA, Janne PA, Mermel C, Pearlberg J, Mukohara T, Fleet C, et al. ErbB-3 mediates phosphoinositide 3-kinase activity in gefitinib-sensitive non-small cell lung cancer cell lines. Proc Natl Acad Sci U S A. 2005;102:3788–93.
Hrustanovic G, Olivas V, Pazarentzos E, Tulpule A, Asthana S, Blakely CM, et al. RAS-MAPK dependence underlies a rational polytherapy strategy in EML4-ALK-positive lung cancer. Nat Med. 2015;21:1038–47.
Blakely CM, Pazarentzos E, Olivas V, Asthana S, Yan JJ, Tan I, et al. NF-kappaB-activating complex engaged in response to EGFR oncogene inhibition drives tumor cell survival and residual disease in lung cancer. Cell Rep. 2015;11:98–110.
Li D, Ambrogio L, Shimamura T, Kubo S, Takahashi M, Chirieac LR, et al. BIBW2992, an irreversible EGFR/HER2 inhibitor highly effective in preclinical lung cancer models. Oncogene. 2008;27:4702–11.
Kwak EL, Sordella R, Bell DW, Godin-Heymann N, Okimoto RA, Brannigan BW, et al. Irreversible inhibitors of the EGF receptor may circumvent acquired resistance to gefitinib. Proc Natl Acad Sci U S A. 2005;102:7665–70.
Finlay MR, Anderton M, Ashton S, Ballard P, Bethel PA, Box MR, et al. Discovery of a potent and selective EGFR inhibitor (AZD9291) of both sensitizing and T790M resistance mutations that spares the wild type form of the receptor. J Med Chem. 2014;57:8249–67.
Soria JC, Ohe Y, Vansteenkiste J, Reungwetwattana T, Chewaskulyong B, Lee KH, et al. Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N Engl J Med. 2018;378:113–25.
Janjigian YY, Smit EF, Groen HJ, Horn L, Gettinger S, Camidge DR, et al. Dual inhibition of EGFR with afatinib and cetuximab in kinase inhibitor-resistant EGFR-mutant lung cancer with and without T790M mutations. Cancer Discov. 2014;4:1036–45.
Acknowledgements
The authors would like to thank Jean-Nicolas Gallant, David Westover, and Karinna Almodovar for their insightful comments during review of this manuscript.
Funding
CML is supported by a Damon Runyon Clinical Investigator Award, LUNGevity Career Development Award, V Foundation Scholar-in-Training Award, AACR-Genentech Career Development Award, LCFA/IASLC Lori Monroe Scholarship, Vanderbilt Ingram Cancer Center Young Ambassadors Award, and by the National Institutes of Health (NIH) and National Cancer Institute (NCI) R01CA121210, P01CA129243, U10CA180864, and P30CA068485-13S5.
Availability of data and materials
Not applicable
Author information
Authors and Affiliations
Contributions
CL conceived the manuscript outline. ZD wrote the initial draft and CL edited. Both authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
ZD reports no potential conflicts of interest. CML has served as a consultant for Pfizer, Novartis, Astra Zeneca, Genoptix, Sequenom, ARIAD, Takeda, and Foundation Medicine and has been an invited speaker for Abbott and Qiagen.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Du, Z., Lovly, C.M. Mechanisms of receptor tyrosine kinase activation in cancer. Mol Cancer 17, 58 (2018). https://doi.org/10.1186/s12943-018-0782-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12943-018-0782-4