Abstract
A plant-associated member of the family Enterobacteriaceae, Serratia plymuthica strain AS12 was isolated from rapeseed roots. It is of scientific interest because it promotes plant growth and inhibits plant pathogens. The genome of S. plymuthica AS12 comprises a 5,443,009 bp long circular chromosome, which consists of 4,952 protein-coding genes, 87 tRNA genes and 7 rRNA operons. This genome was sequenced within the 2010 DOE-JGI Community Sequencing Program (CSP2010) as part of the project entitled “Genomics of four rapeseed plant growth promoting bacteria with antagonistic effect on plant pathogens”.
Similar content being viewed by others
Introduction
Plant associated Serratia species are commonly found as free-living bacteria in rhizosphere soil and as endophytes within plant roots. They include strains with the ability to stimulate plant growth and to inhibit the growth of soil borne pathogens of economically important agricultural plants [1–3]. One Serratia strain, S. plymuthica HRO-C48, is successfully used as an alternative to chemical agents for control of soil-borne fungal diseases in different crops such as strawberry and rapeseed [3,4]. Its ability to degrade chitin, a fungal cell wall component, may be responsible for antifungal activity, whereas the production of the plant hormone indole-3-acetic acid (IAA) could be involved in plant growth promotion [3]. S. plymuthica AS12 has chitinolytic activity and was isolated from rapeseed roots from Uppsala, Sweden in 1998 [5]. The reason for our interest in S. plymuthica AS12 is its ability to inhibit Verticillium longisporum (earlier V. dahliae), a soil borne fungal pathogen of rapeseed, thus promoting the rapeseed growth both directly and indirectly [5]. Here we present a description of the complete genome of S. plymuthica AS12 and its annotation.
Classification and features
A representative 16S rRNA gene sequence of the strain AS12 genome was used for comparison using NCBI BLAST [6] under default settings with the most recent databases. The relative frequencies of taxa and BLAST scores were determined. The most frequently occurring genus is Serratia where some of the ‘hits’ share a 100% identity. When considering high-scoring segment pairs (HSPs) from the best 250 hits, the most frequent matches were Serratia sp. (17.2%) with a maximum identity of 97–100%, while S. plymuthica (5.2%) had a maximum identity of 97–100%, S. proteomaculans (4.8%) with a maximum identity of 97–99%, S. marcescens (4.8%) with a maximum identity of 96–97% and different strains of Rahnella (7%) with a maximum identity of 97–98%.
A phylogenetic tree (Figure 1) was constructed using 16S rRNA sequences of S. plymuthica AS12 with other genera within the family Enterobacteriaceae including two species within the genus Serratia. The tree shows the position of S. plymuthica AS12 within the genus Serratia and its distinct clustering with S. plymuthica, which was confirmed by digital DNA-DNA hybridization values [11] above 70% with the (unpublished) draft genome sequence of the S. plymuthica type strain Breed K-7T from a DSM 4540 culture as well as with the complete genome sequence of S. plymuthica AS9 [12] using the GGDC web server [13].
Phylogenetic tree highlighting the position of S. plymuthica AS12 in relation to selected Serratia strains and other genera within the family Enterobacteriaceae. The tree was based on 1,535 characters of the 16S rRNA gene sequence aligned in ClustalW2 [7]. The tree was inferred under the maximum likelihood criterion using MEGA5 software [8] and rooted with Pseudomonas trivialis (a member of the Pseudomonadaceae family). The branches are mapped by the expected number of substitutions per site. The numbers above the branches are support values from 1,000 bootstrap replicates if larger than 60% [9]. Lineages with genome sequences registered in GOLD [10] are shown in blue.
The cells of strain AS12 stain Gram-negative and are rod shaped, 1–2 µm long, 0.5–0.7 µm wide (Figure 2 and Table 1) and motile. The culture forms red to pink colored colonies of 1–2 mm diameter on tryptic soy agar and potato dextrose agar, but the colony color depends on different factors such as the growth substrate, pH of the medium and growth temperature. The organism is a facultative anaerobe and grows at temperatures ranging from 4 °C – 40 °C and within a pH range of 4–10. It has the ability to utilize a wide range of carbon sources such as glucose, sucrose, succinate, mannitol and arabinose. It also has cellulolytic, phospholytic, chitinolytic and proteolytic activity [5]. The strain is deposited in the Culture Collection, University of Göteborg, Sweden (CCUG) as Serratia sp. AS12 (= CCUG 61397).
Chemotaxonomy
The cells of S. plymuthica AS12 contain a mixture of saturated and unsaturated fatty acids. The dominant fatty acids in strain AS12 are C16:0 (22.94%), C16:1ω7c (17.08%), C18:1ω7c (19.65%), C14:0 (5.11%), along with other minor fatty acid components. No information is available for other compounds. Previously it has been shown that Serratia spp. contain a mixture of C14:0, C16:0, C16:1 and C18:1+2 fatty acids in which 50–80% of the total fatty acid in the cell is C14:0 and others each less than 3% [28]. This is consistent with the observation that C14:0 is a characteristic fatty acid of the family Enterobacteriaceae.
Genome sequencing information
S. plymuthica AS12 was selected for sequencing on the basis of its ability to promote rapeseed plant growth as well as to inhibit fungal pathogens of rapeseed [5]. The genome sequence is deposited in the Genomes On Line Database [10] (GOLD ID = Gc01771) and in GenBank (INSDC ID = CP002774). Sequencing, finishing and annotation were performed by the DOE Joint Genome Institute (JGI). A summary of the project information and its association with MIGS identifiers is shown in Table 2.
Growth conditions and DNA isolation
The cells of S. plymuthica AS12 were grown in Luria Broth (LB) medium at 28°C with constant shaking at 200 rpm. The cells were harvested after 12 hours when the cells were in the early stationary phase. The cells were pelleted and resuspended in TE buffer (Sigma Aldrich). The DNA was extracted from the resuspended cells by following the standard CTAB protocol for bacterial genomic DNA isolation, which is available at JGI [29].
Genome sequencing and assembly
The genome of S. plymuthica AS12 was sequenced using a combination of Illumina [30] and 454 sequencing platforms [31]. The detailed information on library construction and sequencing can be found at the JGI website [29]. The sequence data from Illumina GAii (1,800 Mb) were assembled with Velvet [32] and the consensus sequence was computationally shredded into 1.5 kb overlapping fake reads. The sequencing data from 454 pyrosequencing (81.6 Mb) were assembled with Newbler. The initial draft assembly contained 61 contigs in one scaffold and consensus sequences were computationally shredded into 2 kb overlapping fake reads. The 454 Newbler consensus reads, the Illumina Velvet consensus reads and the read pairs in the 454 paired end library were integrated using a software parallel Phrap [33]. Possible mis-assemblies were corrected with gapResolution [29], Dupfinisher [34], or by sequencing cloned bridging PCR fragments with subcloning or transposon bombing (Epicentre Biotechnologies, Madison, WI). The gaps between contigs were closed by editing in the software Consed [35–37], by PCR and by Bubble PCR (J.-F. Chang, unpublished) primer walks. A total of 160 additional reactions was necessary to close gaps and to raise the quality of the finished sequence. The sequence reads from Illumina were used to correct potential base errors and increase consensus quality using the software Polisher developed at JGI [38]. The final assembly is based on 47.4 Mb of 454 draft data which provides an average 8.8 × coverage of the genome and 315 Mb of Illumina draft data which provides an average 59 × coverage of the genome.
Genome annotation
The S. plymuthica AS12 genes were identified using Prodigal [39] as part of the genome annotation pipeline at Oak Ridge National Laboratory (ORNL), Oak Ridge, TN, USA, followed by a round of manual curation using the JGI GenePRIMP pipeline [40]. The predicted CDS were translated and used to search the National Center for Biotechnology Information (NCBI) nonredundant database, Uniport, TIGR-Fam, Pfam, PRIAM, KEGG, COG and InterPro databases. The miscellaneous functions were predicted using tRNAScan-SE [41], RNAmmer [42], TMHMM [43], and signalP [44]. Additional gene prediction analysis and functional annotation was performed within the Integrated Microbial Genomes — Expert Review (IMG-ER) platform developed by the Joint Genome Institute, Walnut Creek, CA, USA [45].
Genome properties
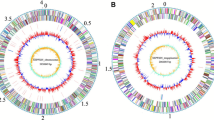
The genome of S. plymuthica AS12 comprises a single circular chromosome of 5,443,009 bp with 55.96% GC content (Figure 3 and Table 3) and 5,140 predicted genes. Among those predicted genes, 4,952 were assigned as protein-coding genes and 88.71% of protein coding genes were assigned for putative function and the remaining ones were annotated as hypothetical proteins. There were 76 pseudogenes and 113 RNA genes with seven rRNA operons. The distribution of genes into the COG functional categories is presented in Table 4.
References
Grimont PAD, Grimont F, Starr MP. Serratia species isolated from plants. Curr Microbiol 1981; 5:317–322. http://dx.doi.org/10.1007/BF01567926
Kalbe C, Marten P, Berg G. Strains of genus Serratia as beneficial rhizobacteria of oilseed rape with antifungal properties. Microbiol Res 1996; 151:433–439. PubMed http://dx.doi.org/10.1016/S0944-5013(96)80014-0
Kurze S, Bahl H, Dahl R, Berg G. Biological control of fungal strawberry diseases by Serratia plymuthica HRO-C48. Plant Dis 2001; 85:529–534. http://dx.doi.org/10.1094/PDIS.2001.85.5.529
Müller H, Berg G. Impact of formulation procedures on the effect of the biocontrol agent Serratia plymuthica HRO-C48 on Verticillium wilt in oilseed rape. BioControl 2008; 53:905–916. http://dx.doi.org/10.1007/s10526-007-9111-3
Alström S. Characteristics of bacteria from oilseed rape in relation to their biocontrol activity against Verticillium dahliae. J Phytopathol 2001; 149:57–64. http://dx.doi.org/10.1046/j.1439-0434.2001.00585.x
Altschul SF, Thomas LS, Alejandro AS, Jingui Z, Webb M, David JL. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res 1997; 25:3389–3402. PubMed http://dx.doi.org/10.1093/nar/25.17.3389
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007; 23:2947–2948. PubMed http://dx.doi.org/10.1093/bioinformatics/btm404
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol 2011; 28:2731–2739. PubMed http://dx.doi.org/10.1093/molbev/msr121
Pattengale ND, Alipour M, Bininda-Emonds ORP, Moret BME, Stamatakis A. How many bootstrap replicates are necessary? Lect Notes Comput Sci 2009; 5541:184–200. http://dx.doi.org/10.1007/978-3-642-02008-7_13
Liolios K, Chen IM, Mavromatis K, Tavernarakis N, Hugenholtz P, Markowitz VM, Kyrpides NC. The Genomes On Line Database GOLD) in 2009: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res 2010; 38:D346–D354. PubMed http://dx.doi.org/10.1093/nar/gkp848
Auch AF, von Jan M, Klenk HP, Göker M. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand Genomic Sci 2010; 2:117–134. PubMed http://dx.doi.org/10.4056/sigs.531120
Neupane S, Högberg N, Alström S, Lucas S, Han J, Lapidus A, Cheng JF, Bruce D, Goodwin L, Pitluck S, et al. Complete genome sequence of the rapeseed plant-growth promoting Serratia plymuthica strain AS9. Stand Genomic Sci 2012; 6:54–62.
Auch AF, Klenk HP, Göker M. Standard operating procedure for calculating genome-to-genome distances based on high-scoring segment pairs. Stand Genomic Sci 2010; 2:142–148. PubMed http://dx.doi.org/10.4056/sigs.541628
Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence MIGS) specification. Nat Biotechnol 2008; 26:541–547. PubMed http://dx.doi.org/10.1038/nbt1360
Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea Bacteria and Eucarya. Proc Natl Acad Sci USA 1990; 87:4576–4579. PubMed http://dx.doi.org/10.1073/pnas.87.12.4576
Garrity GM, Bell JA, Lilburn T. Phylum XIV. Proteobacteria phyl. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT (eds), Bergey’s Manual of Systematic Bacteriology, Second Edition, Volume 2, Part B, Springer, New York, 2005, p. 1.
List Editor. Validation of publication of new names and new combinations previously effectively published outside the IJSEM. List no. 106. Int J Syst Evol Microbiol 2005; 55:2235–2238. http://dx.doi.org/10.1099/ijs.0.64108-0
Garrity GM, Bell JA, Lilburn T. Class III. Gammaproteobacteria class. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT (eds), Bergey’s Manual of Systematic Bacteriology, Second Edition, Volume 2, Part B, Springer, New York, 2005, p. 1.
Garrity GM, Holt JG. Taxonomic Outline of the Archaea and Bacteria. In: Garrity GM, Boone DR, Castenholz RW (eds), Bergey’s Manual of Systematic Bacteriology, Second Edition, Volume 1, Springer, New York, 2001, p. 155–166.
Skerman VBD, McGowan V, Sneath PHA. Approved Lists of Bacterial Names. Int J Syst Bacteriol 1980; 30:225–420. http://dx.doi.org/10.1099/00207713-30-1-225
Rahn O. New principles for the classification of bacteria. Zentralbl Bakteriol Parasitenkd Infektionskr Hyg 1937; 96:273–286.
Judicial Commission. Conservation of the family name Enterobacteriaceae, of the name of the type genus, and designation of the type species OPINION NO. 15. Int Bull Bacteriol Nomencl Taxon 1958; 8:73–74. http://dx.doi.org/10.1099/0096266X-8-1-73
Sakazaki R. Genus IX. Serratia Bizio 1823, 288. In: Buchanan RE, Gibbons NE (eds), Bergey’s Manual of Determinative Bacteriology, Eighth Edition, The Williams and Wilkins Co., Baltimore, 1974, p. 326–326.
Bizio B. Lettera di Bartolomeo Bizio al chiarissimo canonico Angelo Bellani sopra il fenomeno della polenta porporina. Biblioteca Italiana o sia Giornale di Letteratura. [Anno VIII]. Scienze e Arti 1823; 30:275–295.
Breed RS, Murray EGD, Hitchens AP. In: Breed RS, Murray EGD, Hitchens AP (eds), Bergey’s Manual of Determinative Bacteriology, Sixth Edition, The Williams and Wilkins Co., Baltimore, 1948, p. 481–482.
BAuA. 2010, Classification of bacteria and archaea in risk groups. http://www.baua.de TRBA 466, p. 200.
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene Ontology: tool for the unification of biology. Nat Genet 2000; 25:25–29. PubMed http://dx.doi.org/10.1038/75556
Bergan T, Grimont AD, Grimont F. Fatty acids of Serratia determined by gas chromatography. Curr Microbiol 1983; 8:7–11. http://dx.doi.org/10.1007/BF01567306
DOE Joint Genome Institute. http://www.jgi.doe.gov/.
Bennett S. Solexa Ltd. Pharmacogenomics 2004; 5:433–438. PubMed http://dx.doi.org/10.1517/14622416.5.4.433
Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature 2005; 437:326–327. PubMed
Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 2008; 18:821–829. PubMed http://dx.doi.org/10.1101/gr.074492.107
Phrap and Phred for Windows. MacOS, Linux, and Unix. http://www.phrap.com.
Han C, Chain P. 2006. Finishing repeat regions automatically with Dupfinisher. In: Proceeding of the 2006 international conference on bioinformatics & computational biology. Arabina HR, Valafar H (eds), CSREA Press. June 26–29, 2006:141–146.
Ewing B, Green P. Base-Calling of automated sequencer traces using Phred. II. error probabilities. Genome Res 1998; 8:186–194. PubMed
Ewing B, Hillier L, Wendl MC, Green P. Base-Calling of automated sequencer traces using Phred. I. accuracy assessment. Genome Res 1998; 8:175–185. PubMed
Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res 1998; 8:195–202. PubMed
Lapidus A, LaButti K, Foster B, Lowry S, Trong S, Goltsman E. POLISHER: An effective tool for using ultra short reads in microbial genome assembly and finishing. AGBT, Marco Island, FL, 2008.
Hyatt D, Chen GL, LoCascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 2010; 11:119. PubMed http://dx.doi.org/10.1186/1471-2105-11-119
Pati A, Ivanova NN, Mikhailova N, Ovchinnikova G, Hooper SD, Lykidis A, Kyrpides NC. GenePRIMP: a gene prediction improvement pipeline for prokaryotic genomes. Nat Methods 2010; 7:455–457. PubMed http://dx.doi.org/10.1038/nmeth.1457
Schattner P, Brooks AN, Lowe TM. The tRNA scan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res 2005; 33:W686–W689. PubMed http://dx.doi.org/10.1093/nar/gki366
Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 2007; 35:3100–3108. PubMed http://dx.doi.org/10.1093/nar/gkm160
Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 2001; 305:567–580. PubMed http://dx.doi.org/10.1006/jmbi.2000.4315
Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 2004; 340:783–795. PubMed http://dx.doi.org/10.1016/j.jmb.2004.05.028
Markowitz VM, Mavromatis K, Ivanova NN, Chen IMA, Chu K, Kyrpides NC. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics 2009; 25:2271–2278. PubMed http://dx.doi.org/10.1093/bioinformatics/btp393
Acknowledgements
We gratefully acknowledge the help of Elke Lang for providing a culture of reference bacterial strains, Evelyne-Marie Brambilla for extraction of DNA, and Anne Fiebig for assembly of the reference genomes required for digital DNA-DNA (all at DSMZ). The work was conducted by the U.S. Department of Energy Joint Genome Institute is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
Author information
Authors and Affiliations
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Neupane, S., Finlay, R.D., Alström, S. et al. Complete genome sequence of Serratia plymuthica strain AS12. Stand in Genomic Sci 6, 165–173 (2012). https://doi.org/10.4056/sigs.2705996
Published:
Issue Date:
DOI: https://doi.org/10.4056/sigs.2705996