Abstract
Background
Root-knot nematodes (RKNs) cause severe losses in kiwifruit-growing regions. The endophytic bacteria could be biological agents for nematodes management. The aim of this study was the isolation and identification of endophytic bacteria from kiwifruit orchards and the evaluation of their antagonistic ability against RKN in greenhouse conditions.
Results
In this study, the population of nematode and the bacterial strains were isolated from kiwifruit roots and leaves in the Mazandaran and Guilan provinces of Iran. Molecular experiments were conducted to identify and confirm the bacterial isolates and RKN species. Also, the effects of bacterial isolates on nematode reproduction factors (number of galls, egg masses, and second-stage juveniles, J2) and growth parameters of kiwifruit plants were determined. The RKN was confirmed as Meloidogyne incognita by molecular identification. Also, the endophytic bacteria were identified based on supplementary experiments and molecular analyses. A total of 31 bacterial endophytes were identified to be including 12 genera of Bacillus, Pseudomonas, Staphylococcus, Exiguobacterium, Sphingomonas, Agrobacterium, Variovorax, Pantoea, Microbacterium, Streptomyces, Chryseobacterium, and Chitinophaga. Generally, Bacillus and Pseudomonas were the dominant genera that included 29.03 and 22.58% of total isolated bacteria, respectively. In vitro screening assays, P. ananatis 121.en and P. chlororaphis 54.en displayed considerable antagonistic ability on J2 mortality of M. incognita and were selected for greenhouse surveys. The isolates displayed a significant reduction in the number of galls and egg masses on roots and juvenile’s population in pot soil. Moreover, 121.en and 54.en strains significantly increased growth parameters including root fresh weight and shoot fresh weight than the control kiwifruit seedlings.
Conclusions
The bacterial endophytes are safe and have a low risk of managing the RKNs and can be effective microbial bio-fertilizers for improving kiwifruit plant growth under RKNs infections.
Similar content being viewed by others
Background
Kiwifruit (family: Actinidiaceae and genus: Actinidia Lindl) is one of the important horticultural crops in North Iran (Mazandaran and Guilan provinces). Kiwifruit is rich in vitamin C, folic acid, antioxidant properties, high minerals and fiber, and medical applications (Pan et al. 2020).
Root-knot nematodes (RKNs) cause extensive losses to a wide range of economical crops including Actinidia spp. in the world. The nematode causes gall at the root systems and impairs the absorption of water and minerals, which results in wilting, growth suppression, reduction in yield, and eventually death (Xia et al. 2019). The application of non-host plants, rotation, resistant plants, flooding, and nematicides is methods for the management of plant-parasitic nematodes. Recently, beneficial biocontrol agents such as bacteria and fungi were used for nematode’s management (Tran et al. 2019). In recent years, the control of nematodes using biocontrol agents such as endophytic bacteria has increased. These endophytic bacteria are present in the inner tissues of plants, protect the plants against pathogens, affect plant growth by production of secondary metabolites, phytohormone, antibiotics, and siderophores, and trigger the plant defense response (Rat et al. 2021). Nowadays, endophytic bacteria have been used as bio-fertilizers and biocontrol agents which reduce the negative effects of chemical material on the environment and human health (Vetrivelkalai 2019).
The diversity of endophytic bacteria demonstrates that these microorganisms can relate to different plant species (Rat et al. 2021). In previous studies, endophytic bacteria were isolated and identified on the basis of 16S rRNA gene sequence from different plant species. Furthermore, the 16S rRNA gene analysis identified phyla of Proteobacteria, Actinobacteria, Firmicutes, and Bacteroidetes that genera of Pseudomonas, Pantoea, Enterobacter, Stenotrophomonas, Acinetobacter, Serratia (Gammaproteobacteria), Microbacterium, Staphylococcus (Firmicutes), Streptomyces, Arthrobacter (Actinobacteria), Bacillus, Paenibacillus, and Mycobacterium were the abundant genera (Hardoim et al. 2015). It has been reported that the frequency of four phyla of Actinobacteria and Firmicutes at the rate of 34%, Bacteroides, alpha and beta Proteobacteria about 5% was distributed in Pinus species. Also, the genera of Bacillus, Pseudomonas, and Microbacterium are reported from pinus, watermelon, tomato, sweet corn, and pepper (Ponpandian et al. 2019). Furthermore, bacterial endophytes of Variovorax boronicumulans and Agrobacterium tumefaciens were isolated from Phalaris arundinacea L. (Węgrzyn and Felis 2018). The endophytes of V. boronicumulans, Bacillus cereus, and Bacillus aryabhattai are isolated from Lavandula dentata L. (Pereira et al. 2016). Moreover, Pantoea spp. and Bacillus spp. are bacterial endophytes that have been isolated from rice, maize, tomato, and medicinal plants (Abo-Elyousr and Hassan 2021). The Pantoea spp. was the dominant bacterial endophyte in Pellaea calomelanos (Sw.) Link (Mahlangu and Serepa-Dlamini 2018). According to the report of Kim et al. (2019), Proteobacteria classes (α, β, and γ) were the most dominant groups in kiwifruit endophytic communities. Also, Cho et al. (2018) reported that the endophytic Pseudomonas is the dominant genus in kiwifruit.
Moreover, there are many reports about the application of bacterial endophytes as biological control agents (Vetrivelkalai 2019). For instance, it has been reported that endophytic Bacillus amyloliquefaciens decreased decay rate (73.12%) against Botryosphaeria dothidea in kiwifruit (Pang et al. 2021). Bacillus halotolerans LYSX1 has an antagonistic ability for the biocontrol of Meloidogyne javanica (Treub 1885) Chitwood 1949 in Lycopersicon esculentum cv. Sufen (Xia et al. 2019). Streptomyces sp. CBG9 has significant nematicidal ability against Meloidogyne incognita (Kofoid and White 1919) Chitwood 1949 in Coffea canephora Pierre ex A. Froehn (Hoang et al. 2020). Although there are studies on endophytic microorganisms in a wide variety of plants, only limited information is available from endophytic bacteria in kiwifruit. Therefore, the aim of this study was the isolation and identification of endophytic bacteria communities from kiwifruit leaves and roots and the evaluation of their antagonistic potential against M. incognita.
Methods
Preparation and identification of RKNs
The population of RKNs was collected from kiwifruit’s roots in the north of Iran, Mazandaran Province. The single egg masses were propagated on roots of susceptible tomato (Early Urbana variety) in a greenhouse (temperature of 25 °C ± 2 and RH of 70%). After 50 days, the pure nematode population was extracted from tomato roots. Molecular identification of nematode species was performed using species-specific primers of SEC-1F (5'GGGCAAGTAAGGATGCTCTG3')/SEC-1R (5'GCACCTCTTTCATAGCCACG3') (Tesařová et al. 2003) for M. incognita. The DNA extraction was carried out using a modified method of Silva et al. (2000). The polymerase chain reaction (PCR) was done in 50 μl solutions containing 25 μl of 2 × master mix, 1 μl of each primer (10 μm), and 50 ng DNA template. The reaction conditions include 3 min at 94 °C, 35 cycles consisting of 30 s, 94 °C; 30 s, 56 °C; and 1 min, 72 °C, with a final extension of 7 min at 72 °C. The PCR products were separated on an agarose gel (1%) and visualized under UV light.
Plant sampling, isolation, and identification of endophytic bacteria
A total of 200 plant samples (leaf and root) were collected from kiwifruit vineyards in the north of Iran both Mazandaran and Guilan provinces. The samples were transferred immediately to the laboratory of citrus and subtropical fruits research center, Agricultural Research Education and Extension Organization (AREEO), Ramsar, Iran.
Isolation of endophytes was done according to Wicaksono et al. (2018) method. The samples were sterilized with 96% ethanol for 10 s, 2% sodium hypochlorite solution for 3 min followed by washing 3 times (one min each time) with sterile distilled water in a laminar flow cabinet. The disinfected tissues were fragmented in sterilized water for 30–40 min and 30 µL of suspension was cultured on sucrose nutrient agar (NAS) medium after serial dilution. Also, 100 µl of the last wash was transferred to Luria–Bertani (LB) as a control (Taechowisan et al. 2003). The plates were incubated at 25 °C for 7 days. The colonies were re-cultured on NAS plates until pure colonies were obtained. Single colonies were stored in 60% glycerol and were stored at − 80 °C for future studies.
A total of 100 isolates were grouped based on morphological features such as filamentous shapes, rough and smooth texture, flat and irregular-edged, and white, yellow, yellow to orange, cream, lemon yellow, and orange colors and gram reaction. For molecular identification, 31 colonies with morphological differences were selected. Bacterial genomic DNA was extracted using the cells lysis protocol (Keegan et al. 2005). The extracted DNA was used for the amplification of the 16S rRNA gene with the universal bacterial primers FD1 (5'AGAGTTTGATCCTGGCTCAG3') and RP2 (5'ACGGTTACCTTGTTACGACTT3') (Weisburg et al. 1991). PCR was performed in 50 μl reactions containing 50 ng DNA template, 1 μl of each primer (10 pmol × μl−1), 25 μl Taq DNA Polymerase 2 × Master Mix RED (Ampliqon, Cat. No. A180301) using a MJ RESEARCH PTC-200 thermal cycler. The amplification program contains initial denaturation at 94 °C for 5 min, 30 cycles of denaturation at 94 °C for 50 s, annealing at 55 °C for 45 s, and extension at 72 °C for 90 s, and a final extension for 3 min at 72 °C. The PCR products (~ 1500 bp) were separated using agarose gel 1% with TBE buffer 1X, stained with SinaClon DNA safe Stain (Cat. No. EP5082, Co., Tehran, Iran), and visualized under UV light. The amplicons were sequenced and the results were compared to the sequences deposited in the national center for biotechnology information database (www.ncbi.nlm.nih.gov). The obtained sequences were aligned using the multiple sequence alignment in MEGA X software. Phylogenetic analysis was carried out using the maximum likelihood method with bootstrapping (1000 replications). Then sequences were deposited in GenBank for obtaining accession numbers.
In vitro nematicidal activity
For evaluation of antagonistic effects of endophytic isolates on mortality of J2s, the suspension of freshly hatched juveniles was used. The isolated eggs from the tomato roots were incubated at 27 ± 2 °C, and freshly hatched juveniles were used for the experiment.
Anti-nematode activity: One ml suspension of selected bacterial isolates (108 CFU/ml) was added to 1 ml of 50 J2s and kept at room temperature. Nematode (J2) mortality was recorded at 48 and 72 h after treatment under a stereoscopic microscope with a magnification of 4×. The negative control with sterile water was performed under the same conditions. Each treatment included five replications and the experiment was repeated thrice. The number of dead juveniles was counted 1–3 days and the mortality ratio of J2s was calculated.
Greenhouse experiments
The endophytic bacterial isolates (121.en and 54.en) with the highest nematicidal activity in the in vitro assays were selected for the greenhouse experiments. For this purpose, 40 ml of the bacterial suspension (1 × 108 CFU/ ml) were treated to six-month-old seedlings of Actinidia chinensis var. deliciosa (A. Chev) cv. Hayward and after 2 days, seedlings were inoculated with 2000 J2s of M. incognita and kept in a greenhouse at 25 ± 4 °C. The treatments were including the seedlings treated only with sterile water (Control); inoculated only with nematode; inoculated with nematode and treated with 121.en; and inoculated with nematode and treated with 54.en. The experiment was carried out in a randomized block design with five replications and repeated thrice. After 50 days, the number of galls and egg masses, fresh and dry weights of shoot and root, and juvenile population of nematodes in the soil were measured.
Statistical analysis
Data were analyzed using SAS version 9.1 software with a one-way variance analysis (ANOVA) test. The mean ± standard deviation (X ± SD) was expressed in all experimental data. The significance of differences (P < 0.05) within treatments was determined using Duncan’s test.
Results
Identification of M. incognita
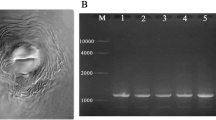
PCR was performed using specific primers of Inc-k14 and fragments of about 502 bp were produced for M. incognita (Fig. 1).
Identification of endophytic bacteria from kiwifruit
The phylogenetic analysis grouped the isolates into six groups and 12 genera (Bacillus, Pseudomonas, Chryseobacterium, Microbacterium, Pantoea, Streptomyces, Sphingomonas, Agrobacterium, Chitinophaga, Staphylococcus, Variovorax, and Exiguobacterium) (Table 1 and Fig. 2). The accession numbers and relationships between genera or species of these endophytic bacteria with sequences deposited in the GenBank database are demonstrated in Table 1. All of the consensus sequences obtained in this study presented a high similarity (up to 97%) with the closest related sequences deposited in the GenBank. Phylogenetic analysis of these strains is demonstrated in Fig. 2.
Phylogenetic tree of isolated endophytic bacteria from kiwifruit based on the 16 s rRNA gene sequences constructed in the MEGA X program based on the maximum likelihood method and bootstrap values (values > 50%) based on 1000 replications. Bacillus megaterium and Staphylococcus sciuri have changed to Priestia megaterium and Mammaliicoccus sciuri
Results allowed us to classify the isolated kiwifruit endophytic bacteria into 6 distinct groups. Group I was belonging to the Firmicutes phylum. This group includes isolates that belonged to the Bacillus (123.en, 60.en, 1.en, 10.en, 31.en, Q.en, M.en, N.en, and K.en), Staphylococcus (27.en and X.en) Exiguobacterium (O.en), which was the most dominant groups in this study. Group II (α-Proteobacteria phylum) including Agrobacterium (isolates 2.en and 19.en) and Sphingomonas (isolate 5.en), Group III (β-Proteobacteria phylum), two bacteria belonged to the Variovorax (isolates 48.en and 53.en), Group IV (Gammaproteobacteria phylum), seven bacteria belonged to the Pseudomonas (isolates 54.en, 41.en, 14.en, 24.en 28.en, 51.en, and P.en) and one bacterium belonged to the Pantoea (isolates 121.en), Group V (Actinobacteria phylum), two bacteria belonged to the Microbacterium (isolates 3.en and 6.en), and one bacteria belonged to the Streptomyces (isolates J.en), and Group VI (Bacteroidetes phylum) contained two bacteria belonged to the Chryseobacterium (isolates 7.en and 44.en) and one bacteria belonged to the Chitinophaga (43.en) (Fig. 2).
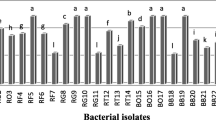
Among these groups, 12 isolates (29.03%) were gram-positive, rod shapes, spore-forming, and irregular-edged belonging to the Bacillus genus. Furthermore, seven isolates (22.58%) were gram-negative, motile, non-spore-forming, catalase-positive, and oxidase negative identified as Pseudomonas spp. Thus, according to the observed results, Bacillus and Pseudomonas were the most dominant genera among isolates and the other genera had a frequency of between 3 and 10% (Fig. 3).
In vitro activity
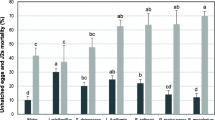
Statistical analyses indicated significant differences among bacterial isolates in terms of J2s mortality. The results of some isolates are given in Table 2. The mean comparison of isolates data indicated that P. ananatis 121.en and P. chlororaphis 54.en had the most effect on J2s mortality and increased the percentage of J2 mortality to 83.3 and 79.26% after 72 h., respectively (Table 2). Thus, these two isolates were selected for greenhouse experiments.
Greenhouse experiment
Two isolates of P. ananatis 121.en and P. chlororaphis 54.en were selected for greenhouse experiments based on the high antagonistic ability on J2s mortality in vitro. Results of the greenhouse experiments demonstrated that endophytic bacteria had positive effects in kiwifruit seedlings inoculated with M. incognita. Indeed, there were significant differences among treatments in nematode reproductive parameters and plant growth parameters in comparison with control plants. The 121.en and 54.en isolates were able to reduce the number of galls and egg masses in the roots of kiwifruit seedlings infected with the nematode. The reduction in the number of galls was 75.11% (121.en) and 74.83% (54.en), and the reduction in the number of egg masses was 86.63% (121.en) and 86.80% (54.en). Also, those were able to reduce the J2s population in the soil, formed by 92% (121.en), and 92.59% (54.en) (Fig. 4). Furthermore, seedlings treated with these three isolates promoted kiwifruit plant growth including the root and shoot fresh weight and subsequently root and shoot dry weight as compared to positive control seedlings (Table 3).
Discussion
This study focused on the diversity of endophytic bacteria and the evaluation of biological control efficacy against M. incognita. A few plants have been studied for their endophytic organisms (Sharma et al. 2018). There are many reports on the isolation of endophytic bacteria from kiwifruit. It has been founded that Pseudomonas (Xie et al. 2021) and Stenotrophomonas (Cho et al. 2018) were the dominant genera in kiwifruit. Some studies have shown the antagonistic ability of endophytic bacteria against plant pathogens in kiwifruit. The endophytes including Pseudomonas fluorescens, P. putida, P. mendocina, Pantoea agglomerans, and Kluyvera intermedia had inhibitory ability against P. syringae pv. actinidiae in kiwifruit (Tontou et al. 2016b). Also, the P. synxantha was effective against P. syringae pv. actinidiae in Actinidia chinensis (Tontou et al. 2016a). Previous findings also confirmed the great effects of endophytic bacteria on controlling nematodes. For example, it was reported that B. megaterium DS9 reduced the nematode population and affected pepper growth promotion (Tran et al. 2019). In the other research, endophytic Streptomyces sp. isolated from banana had high antagonistic potential against M. javanica (Su et al. 2017). Vetrivelkalai (2019) reported that isolates of Bacillus sp. and Pseudomonas sp. reduced the number of egg masses and M. incognita population in the soil. In another research, Sharma and Sharma (2017) reported Pseudomonas sp. reduced the nematode infection and increased the growth parameters in tomato plants. Also, Khanna et al. (2019) revealed that P. aeruginosa increased root dry weight and fresh weight 17.5 and 16.4% in Lycopersicon esculentum Mill. seedlings. Indeed, the results of this study were in agreement with these reports that 121.en and 54.en isolates significantly reduced the number of galls and egg masses and promoted plant growth in greenhouse assays.
The 16S rRNA gene sequencing is important for phylogeny studies and is a useful tool for the identification of bacterial endophytes to subspecies level in a variety of plants ((Liaqat and Eltem 2016). The results of the present study showed that the most dominant genus was Bacillus, with a frequency of 29.03%, which was not organ-specific and isolated from leaf and root tissues, and the strains of B. altitudinis 123.en and 10.en, B. safensis Q.en, B. megaterium 31.en and 60.en, B. halotolerans 1.en and Bacillus sp. K.en, M.en, and N.en, reported from the Bacillus genus. The Bacillus genus is an endophytic bacterium from the phylum Firmicutes and the family of Bacillaceae. The Bacillus species are important due to auxin, gibberellin, and siderophore production, phosphate solubilization, and the ability to adapt to drought (Dias et al. 2009). This genus is effective for the biological control of plant diseases against Meloidogyne hapla Chitwood 1949, Fusarium oxysporum, and other pathogens (Ma et al. 2013). There are reports on the biological control of plant diseases using endophytic bacteria. For example, the B. altitudinis with the production of main metabolites such as cyclic lipopeptides is a proper candidate as a bio-fertilizer and plant growth promoter in sustainable agriculture (Zhang et al. 2021). The B. halotolerans Y6 with overexpressing β-glucanase inhibits the mycelial growth of Verticillium dahlia because the production of secondary metabolites is important in biological control (Zhang et al. 2019). B. safensis ZY16 isolated from Chloris virgata Sw. with the production of biosurfactants and degradation of hydrocarbons causes plant growth promotion (Wu et al. 2019). The endophytic B. megaterium RmBm31 isolated from Retama monosperma (L.) Boiss. via solubilization of phosphate and production of indole-3-acetic acid (IAA) causes plant growth-promoting (Dahmani et al. 2020). The second genus in this study was the Pseudomonas, with a frequency of 29.03% that strains of P. fulva 28.en, P. chlororaphis 14.en, 41.en, 51.en, 54.en, P.en and P. psychrotolerans 24.en, reported from Pseudomonas genus. The Pseudomonas from the class Gammaproteobacteria and the family Pseudomonadaceae are reported in most plants. Pseudomonas genus due to the production of various antibiotics including HCN, 2, 4-diacetylphloroglucinol, and pyrrolnitrin is affected plant growth (Whipps 2001). It has been reported that endophytic P. fulva strain MRC41 isolated from maize with activities of plant growth promotion and drought tolerance helped plant disease management (Sandhya et al. 2017). P. chlororaphis strains due to the production of antibiotics and trigger of systemic resistance protect plants against plant diseases. The endophytic P. psychrotolerans isolated from Taxus chinensis (Rehder & E.H.Wilson) Rehder via the production of antioxidants has antagonistic activity and is effective for plant growth promotion (Fidan and Zhan 2019). The third genus in this study with a frequency of 6.45% was from the Chryseobacterium genus, family Flavobacteriaceae. An endophytic Chryseobacterium sp. GSE06 isolated from Cucumis sativus L. was a biocontrol agent against Phytophthora capsici. This genus has the ability of colonization, plant growth promotion, and antimicrobial activity (Jeong et al. 2016). Eke et al. (2019) reported that the endophytic Ch. indologenes isolated from Euphorbia trigona Mill was effective for tomato growth promotion. The fourth genus, with a frequency of 6.45%, belongs to the genus Microbacterium from the family of Microbacteriaceae that in this study strains of M. foliorum 3.en and 6.en reported from Microbacterium genus. The Microbacterium strains with produce secondary metabolites (antibiotics, solubilize Potassium, pigments, and siderophores) have an important role in biological control (Corretto et al. 2020). For example, the M. foliorum CT10 isolated from tomato stems and leaves was effective against F. oxysporum and Botrytis cinerea (Hernández-Pacheco et al. 2021). The fifth genus was the Staphylococcus, with a frequency of 6.45% that in this study, strains of S. sciuri 27.en and X.en were reported from Staphylococcus genus. They belong to the family Staphylococcaceae that have salt tolerance potential and influence in the release of growth regulators and produce gibberellin, IAA, protease, chitinase, and siderophore (Alijani et al. 2019). The endophytic Staphylococcus sciuri reported from roots Leptochloa fusca L. by phosphate solubilization and production of phytohormone regulates plant development. However, this genus plays the important role in the biological control of plant pathogens (Dutta et al. 2017), for example, S. sciuri MarR44 as an endophytic bacterial isolated from strawberry due to the production of volatile compounds had antifungal activity against strawberry anthracnose (Alijani et al. 2019). The sixth genus was the Variovorax (family of Comamonadaceae), with a frequency of 6.45% that in this study, strains of V. boronicumulans 48.en and 53.en were reported from Variovorax genus. This genus has an important role in microbe–plant interactions. V. boronicumulans regulates hormone IAA levels in plants (Sun et al. 2018). It has been reported that the V. boronicumulans CGMCC4969 promotes plant growth by producing siderophores, hydrogen cyanide, ammonia, and secreting salicylate (Liu et al. 2013). The seventh genus, with a frequency of 3.22%, belongs to the genus Chitinophaga from the family of Chitinophagaceae that in this research, the strain of Chitinophaga sp. 43.en, reported from Chitinophaga genus. This genus produces secondary metabolites with antimicrobial activity and is an effective endophyte for the biological control of pathogens such as Rhizoctonia solani (Carrión et al. 2019). It has been reported that in the Chitinophaga MR33 strain via multifunctional activities such as phosphate solubilization, protease, and chitinolytic activity, IAA production is effective for plant growth promotion (Chimwamurombe et al. 2016). The eighth genus was the Agrobacterium, with a frequency of 6.45% that in this study, strains of A. tumefaciens 2.en and 19.en were reported from Agrobacterium genus. The genus belongs to the family of Rhizobiaceae. There are reports of endophytic A. tumefaciens strains with the potential of plant-growth-promoting that have been isolated from various plants. The endophytic A. tumefaciens CCNWGS0286 isolated from the Robinia pseudoacacia L. had a key role as plant-growth-promoting (Hao et al. 2012). Too, A. tumefaciens CR22 isolated from tomato roots effective against F. oxysporum (Hernández-Pacheco et al. 2021). The ninth genus, with a frequency of 3.22%, belonged to the genus Pantoea from the family of Erwiniaceae. In this study, the strain of P. ananatis 121.en was reported from Pantoea genus. It has been reported that P. ananatis D1 promotes plant growth by producing siderophore, 1-aminocyclopropane-1-carboxylic acid deaminase, and indole-3-acetic acid (Lu et al. 2021). The tenth genus, with a frequency of 3.22%, belongs to the genus Sphingomonas from the family of Sphingomonadaceae. The strain of Sphingomonas sp. 5.en was reported from Sphingomonas genus. The Sphingomonas strains have beneficial properties as endophytes in various plants such as papaya (Rivarez et al. 2021), and Allium tuberosum (Huang 2019). Sphingomonas sp. as a common plant bacterial endophyte is beneficial for plants by increasing of producing phytohormone. The eleventh genus, with a frequency of 3.22%, belongs to the genus Streptomyces from the family of Streptomycetaceae. The strain of Streptomyces sp. J.en reported from Streptomyces genus. Streptomyces bacteria as biocontrol agents produced secondary metabolites such as antimicrobials volatile compounds, IAA hormone, and plant protection from biotic stresses, which colonize plant roots. They stimulate plant growth and control different bacterial and fungal pathogens (Vurukonda et al. 2021). The last genus, with a frequency of 3.22%, belongs to the genus Exiguobacterium from the family of Bacillaceae. The strain of E. acetylicum O.en was reported from Exiguobacterium genus. The Exiguobacterium acetylicum had the potential of phosphate solubilizing, ability of enzyme production, plant growth promotion and increases plant tolerance to cold (Selvakumar et al. 2010). The E. acetylicum was isolated from C4 plants (Girsowicz et al. 2019). It has been reported that E. acetylicum suppressed the growth of the plant pathogens F. oxysporum, Sclerotium rolfsii, R. solani, and Pythium sp (Selvakumar et al. 2010).
Generally, to manage nematodes, two stages of second-stage juvenile and eggs should be controlled because these are important in their life cycle; interruption in these stages results in a decrease in nematode populations (Xiang et al. 2018). Endophytic bacteria by the production of enzymes, HCN, hormones, secondary metabolites, and induction of plant resistance suppress pathogens. These bacteria by producing chitinase, protease, catalase, lipase enzymes, and HCN can kill nematodes. Also, they promote plant growth by dissolving phosphorus and stabilizing nitrogen (Ma et al. 2016). In the present study, isolation of endophytic bacteria was performed from healthy kiwifruit trees in nematodes-infected orchards. In these orchards, some trees were unaffected by nematodes and remained healthy. The findings may show that this can be due to beneficent microorganisms such as endophytes in the plant and rhizosphere. The greenhouse experiments were conducted to assess the biological control efficacy of endophytic bacteria against RKN in kiwifruit seedlings. The results showed that the treatment of endophytic bacteria P. ananatis 121.en and P. chlororaphis 54.en reduced the numbers of galls and egg masses and promoted the growth of kiwifruit plants in compared with control plants. Also, the rate of J2 mortality increased in the soil 50 days after inoculation. Similar results were indicated by Vetrivelkalai (2019) who reported that the number of galls, egg masses, and population of RKNs were significantly reduced and the plant growth parameters were promoted in plants treated with endophytic bacteria of Pseudomonas, Pantoea, and Bacillus. Therefore, obtained results indicated that endophytic bacteria of P. ananatis 121.en and P. chlororaphis 54.en can be potential agents against M. incognita and provide a useful option for the selection of kiwifruit growth-promoting microorganisms.
Conclusions
The results showed that endophytic bacteria isolated from kiwifruit vine had a high diversity genetically and belonged to different phylogenetic groups from genera Bacillus, Pseudomonas, Chryseobacterium, Microbacterium, Pantoea, Streptomyces, Variovorax, and Exiguobacterium. This diversity indicates the fact that kiwifruit vine habitats for a variety of gram-positive and gram-negative endophytic bacteria. Moreover, the results showed that the antagonistic ability of two isolates P. ananatis 121.en and P. chlororaphis 54.en against M. incognita resulted in a reduction in the number of gall and egg masses in the roots, a decrease in the J2 population in the soil, and improvement in plant growth parameters. Thus, these endophytic bacteria can be both plant growth stimulators and biocontrol agents against root-knot nematodes. On the other hand, these isolates can be a good candidate for microbial formulation to manage pathogens and improve plant growth. Therefore, large-scale field experiments should be performed in the future.
Availability of data and materials
All data generated during the current study are included in this article, and sequence data generated are available as nucleotide sequence in the NCBI GenBank.
Abbreviations
- RKNs:
-
Root-knot nematodes
- J2:
-
Second-stage juvenile
- AREEO:
-
Agricultural Research Education and Extension Organization
- NAS:
-
Sucrose nutrient agar
- LB:
-
Luria–Bertani
- PCR:
-
Polymerase chain reaction
- IAA:
-
Indole-3-acetic acid
References
Abo-Elyousr KA, Hassan SA (2021) Biological control of Ralstonia solanacearum (Smith), the causal pathogen of bacterial wilt disease by using Pantoea spp. Egypt J Biol Pest Control 31(1):1–8. https://doi.org/10.1186/s41938-021-00460-z
Alijani Z, Amini J, Ashengroph M, Bahramnejad B (2019) Antifungal activity of volatile compounds produced by Staphylococcus sciuri strain MarR44 and its potential for the biocontrol of Colletotrichum nymphaeae, causal agent strawberry anthracnose. Int J Food Microbiol 307:108276. https://doi.org/10.1016/j.ijfoodmicro.2019.108276
Carrión VJ, Perez-Jaramillo J, Cordovez V, Tracanna V, De Hollander M, Ruiz-Buck D, Mendes LW, Van Ijcken WF, Gomez-Exposito R, Elsayed SS (2019) Pathogen-induced activation of disease-suppressive functions in the endophytic root microbiome. Sci 366(6465):606–612. https://doi.org/10.1126/science.aaw9285
Chimwamurombe PM, Grönemeyer JL, Reinhold-Hurek B (2016) Isolation and characterization of culturable seed-associated bacterial endophytes from gnotobiotically grown Marama bean seedlings. FEMS Microbiol Ecol. https://doi.org/10.1093/femsec/fiw083
Cho G, Kim M-j, Kwon Y, Kwak Y-S (2018) Comparison of endophytic microbial community in kiwifruit plant cultivars. Plant Pathol J 34(4):341. https://doi.org/10.5423/ppj.nt.12.2017.0284
Corretto E, Antonielli L, Sessitsch A, Höfer C, Puschenreiter M, Widhalm S, Swarnalakshmi K, Brader G (2020) Comparative genomics of Microbacterium species to reveal diversity, potential for secondary metabolites and heavy metal resistance. Front in Microbiol 11:1869. https://doi.org/10.3389/fmicb.2020.01869
Dahmani MA, Desrut A, Moumen B, Verdon J, Mermouri L, Kacem M, Coutos-Thévenot P, Kaid-Harche M, Bergès T, Vriet C (2020) Unearthing the plant growth-promoting traits of Bacillus megaterium RmBm31, an endophytic bacterium isolated from root nodules of Retama monosperma. Front Plant Sci 11:124. https://doi.org/10.3389/fpls.2020.00124
Dias AC, Costa FE, Andreote FD, Lacava PT, Teixeira MA, Assumpçao LC, Araújo WL, Azevedo JL, Melo IS (2009) Isolation of micropropagated strawberry endophytic bacteria and assessment of their potential for plant growth promotion. World J Microbiol Biotechnol 25(2):189–195. https://doi.org/10.1007/s11274-008-9878-0
Dutta A, Ghosh S, Choudhury JD, Mahansaria R, Roy M, Ghosh AK, Roychowdhury T, Mukherjee J (2017) Isolation of indigenous Staphylococcus sciuri from chromium-contaminated paddy field and its application for reduction of Cr (VI) in rice plants cultivated in pots. Bioremediat J 21(1):30–37. https://doi.org/10.1080/10889868.2017.1282935
Eke P, Kumar A, Sahu KP, Wakam LN, Sheoran N, Ashajyothi M, Patel A, Fekam FB (2019) Endophytic bacteria of desert cactus (Euphorbia trigonas Mill) confer drought tolerance and induce growth promotion in tomato (Solanum lycopersicum L.). Microbiol Res 228:126302. https://doi.org/10.1016/j.micres.2019.126302
Fidan O, Zhan J (2019) Discovery and engineering of an endophytic Pseudomonas strain from Taxus chinensis for efficient production of zeaxanthin diglucoside. J Biol Eng 13(1):1–18. https://doi.org/10.1186/s13036-019-0196-x
Girsowicz R, Moroenyane I, Steinberger Y (2019) Bacterial seed endophyte community of annual plants modulated by plant photosynthetic pathways. Microbiol Res 223:58–62. https://doi.org/10.1016/j.micres.2019.03.001
Hao X, Xie P, Johnstone L, Miller SJ, Rensing C, Wei G (2012) Genome sequence and mutational analysis of plant-growth-promoting bacterium Agrobacterium tumefaciens CCNWGS0286 isolated from a zinc-lead mine tailing. Appl Environ Microbiol 78(15):5384–5394. https://doi.org/10.1128/AEM.01200-12
Hardoim PR, Van Overbeek LS, Berg G, Pirttilä AM, Compant S, Campisano A, Döring M, Sessitsch A (2015) The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol Mol Biol Rev 79(3):293–320. https://doi.org/10.1128/MMBR.00050-14
Hernández-Pacheco CE, del Carmen Orozco-Mosqueda M, Flores A, Valencia-Cantero E, Santoyo G (2021) Tissue-specific diversity of bacterial endophytes in Mexican husk tomato plants (Physalis ixocarpa Brot ex. Horm.), and screening for their multiple plant growth-promoting activities. Current Res Microb Sci 2:100028. https://doi.org/10.1016/j.crmicr.2021.100028
Hoang H, Tran LH, Nguyen TH, Nguyen DAT, Nguyen HHT, Pham NB, Trinh PQ, de Boer T, Brouwer A, Chu HH (2020) Occurrence of endophytic bacteria in Vietnamese Robusta coffee roots and their effects on plant parasitic nematodes. Symbiosis 80(1):75–84. https://doi.org/10.1007/s13199-019-00649-9
Huang Y (2019) Illumina-based analysis of endophytic bacterial diversity of four Allium species. Sci Rep 9(1):1–11. https://doi.org/10.1038/s41598-019-51707-7
Jeong J-J, Park BH, Park H, Choi I-G, Kim KD (2016) Draft genome sequence of Chryseobacterium sp. strain GSE06, a biocontrol endophytic bacterium isolated from cucumber (Cucumis sativus). Genome Announc 4(3):e00577. https://doi.org/10.1128/genomeA.00577-16
Keegan H, Boland C, Malkin A, Griffin M, Ryan F, Lambkin H (2005) Comparison of DNA extraction from cervical cells collected in PreservCyt solution for the amplification of Chlamydia trachomatis. Cytopathol 16(2):82–87. https://doi.org/10.1111/j.1365-2303.2005.00239.x
Khanna K, Jamwal VL, Kohli SK, Gandhi SG, Ohri P, Bhardwaj R, Wijaya L, Alyemeni MN, Ahmad P (2019) Role of plant growth promoting bacteria (PGPRs) as biocontrol agents of Meloidogyne incognita through improved plant defense of Lycopersicon esculentum. Plant Soil 436(1):325–345. https://doi.org/10.1007/s11104-019-03932-2
Kim M-J, Do H, Cho G, Jeong R-D, Kwak Y-S (2019) Comparison of microbial community of rhizosphere and endosphere in kiwifruit. Plant Pathol J 35(6):705. https://doi.org/10.5423/ppj.nt.08.2019.0216
Liaqat F, Eltem R (2016) Identification and characterization of endophytic bacteria isolated from in vitro cultures of peach and pear rootstocks. 3 Biotech 6(2):1–8. https://doi.org/10.1007/s13205-016-0442-6
Liu Z-H, Cao Y-M, Zhou Q-W, Guo K, Ge F, Hou J-Y, Hu S-Y, Yuan S, Dai Y-J (2013) Acrylamide biodegradation ability and plant growth-promoting properties of Variovorax boronicumulans CGMCC 4969. Biodegradation 24(6):855–864. https://doi.org/10.1007/s10532-013-9633-6
Lu L, Chang M, Han X, Wang Q, Wang J, Yang H, Guan Q, Dai S (2021) Beneficial effects of endophytic Pantoea ananatis with ability to promote rice growth under saline stress. J Appl Microbiol 131(4):1919–1931. https://doi.org/10.1111/jam.15082
Ma L, Cao YH, Cheng MH, Huang Y, Mo MH, Wang Y, Yang JZ, Yang FX (2013) Phylogenetic diversity of bacterial endophytes of Panax notoginseng with antagonistic characteristics towards pathogens of root-rot disease complex. Anton Leeuw 103(2):299–312. https://doi.org/10.1007/s10482-012-9810-3
Ma Y, Rajkumar M, Zhang C, Freitas H (2016) Beneficial role of bacterial endophytes in heavy metal phytoremediation. J Environ Manag 174:14–25. https://doi.org/10.1016/j.jenvman.2016.02.047
Mahlangu SG, Serepa-Dlamini MH (2018) First report of bacterial endophytes from the leaves of Pellaea calomelanos in South Africa. S Afr J Sci 114(9–10):1–9. https://doi.org/10.17159/sajs.2018/4235
Pan L, Zhao X, Chen M, Fu Y, Xiang M, Chen J (2020) Effect of exogenous methyl jasmonate treatment on disease resistance of postharvest kiwifruit. Food Chem 305:125483. https://doi.org/10.1016/j.foodchem.2019.125483
Pang L, Xia B, Liu X, Yi Y, Jiang L, Chen C, Li P, Zhang M, Deng X, Wang R (2021) Improvement of antifungal activity of a culture filtrate of endophytic Bacillus amyloliquefaciens isolated from kiwifruit and its effect on postharvest quality of kiwifruit. J Food Biochem 45(1):e13551. https://doi.org/10.1111/jfbc.13551
Pereira S, Monteiro C, Vega A, Castro PM (2016) Endophytic culturable bacteria colonizing Lavandula dentata L. plants: isolation, characterization and evaluation of their plant growth-promoting activities. Ecol Eng 87:91–97. https://doi.org/10.1016/j.ecoleng.2015.11.033
Ponpandian LN, Rim SO, Shanmugam G, Jeon J, Park Y-H, Lee S-K, Bae H (2019) Phylogenetic characterization of bacterial endophytes from four Pinus species and their nematicidal activity against the pine wood nematode. Sci Rep 9(1):1–11. https://doi.org/10.1038/s41598-019-48745-6
Rat A, Naranjo HD, Krigas N, Grigoriadou K, Maloupa E, Alonso AV, Schneider C, Papageorgiou VP, Assimopoulou AN, Tsafantakis N (2021) Endophytic bacteria from the roots of the medicinal plant Alkanna tinctoria Tausch (Boraginaceae): exploration of plant growth promoting properties and potential role in the production of plant secondary metabolites. Front in Microbiol 12:113. https://doi.org/10.3389/fmicb.2021.633488
Rivarez MPS, Parac EP, Dimasingkil SFM, Mirnia E, Magdalita PM (2021) Influence of native endophytic bacteria on the growth and bacterial crown rot tolerance of papaya (Carica papaya). Eur J Plant Pathol 161(3):593–606. https://doi.org/10.1007/s10658-021-02345-1
Sandhya V, Shrivastava M, Ali SZ, Prasad VSSK (2017) Endophytes from maize with plant growth promotion and biocontrol activity under drought stress. Russ Agricult Sci 43(1):22–34. https://doi.org/10.3103/S1068367417010165
Selvakumar G, Kundu S, Joshi P, Nazim S, Gupta A, Gupta H (2010) Growth promotion of wheat seedlings by Exiguobacterium acetylicum 1P (MTCC 8707) a cold tolerant bacterial strain from the Uttarakhand Himalayas. Indian J Microbiol 50(1):50–56. https://doi.org/10.1007/s12088-009-0024-y
Sharma R, Tangjang S, Wangpan T (2018) First report on biological evaluation and preliminary screening of fungal eendophytes from spilanthes paniculata, a medicinal herb in Arunachal Pradesh, India. Int J Curr Microbiol App Sci 7(11):1346–1354. https://doi.org/10.20546/ijcmas.2018.711.157
Sharma IP, Sharma A (2017) Co-inoculation of tomato with an arbuscular mycorrhizal fungus improves plant immunity and reduces root-knot nematode infection. Rhizosphere 4:25–28. https://doi.org/10.1016/j.rhisph.2017.05.008
Silva ATd, Penna JC, Goulart LR, Santos MAd, Arantes NE (2000) Genetic variability among and within races of Heterodera glycines Ichinohe assessed by RAPD markers. Genet Mol Biol 23:223–229. https://doi.org/10.1590/S1415-47572000000200014
Su L, Shen Z, Ruan Y, Tao C, Chao Y, Li R, Shen Q (2017) Isolation of antagonistic endophytes from banana roots against Meloidogyne javanica and their effects on soil nematode community. Front in Microbiol 8:2070. https://doi.org/10.3389/fmicb.2017.02070
Sun S-L, Yang W-L, Fang W-W, Zhao Y-X, Guo L, Dai Y-J (2018) The plant growth-promoting rhizobacterium Variovorax boronicumulans CGMCC 4969 regulates the level of indole-3-acetic acid synthesized from indole-3-acetonitrile. Appl Environ Microbiol 84(16):e00298. https://doi.org/10.1128/AEM.00298-18
Taechowisan T, Peberdy JF, Lumyong S (2003) Isolation of endophytic actinomycetes from selected plants and their antifungal activity. World J Microbiol Biotechnol 19(4):381–385. https://doi.org/10.1023/A:1023901107182
Tesařová B, Zouhar M, Ryšánek P (2003) Development of PCR for specific determination of root-knot nematode Meloidogyne incognita. Plant Prot Sci 39(1):23. https://doi.org/10.17221/3823-PPS
Tontou R, Gaggia F, Baffoni L, Devescovi G, Venturi V, Giovanardi D, Stefani E (2016a) Molecular characterisation of an endophyte showing a strong antagonistic activity against Pseudomonas syringae pv. actinidiae. Plant Soil 405(1):97–106. https://doi.org/10.1007/s11104-015-2624-0
Tontou R, Giovanardi D, Ferrari M, Stefani E (2016b) Isolation of bacterial endophytes from Actinidia chinensis and preliminary studies on their possible use as antagonists against Pseudomonas syringae pv. actinidiae. J Berry Res 6(4):395–406. https://doi.org/10.3233/JBR-160118
Tran TPH, Wang S-L, Nguyen VB, Tran DM, Nguyen DS, Nguyen AD (2019) Study of novel endophytic bacteria for biocontrol of black pepper root-knot nematodes in the central highlands of Vietnam. J Agron 9(11):714. https://doi.org/10.3390/agronomy9110714
Vetrivelkalai P (2019) Evaluation of endophytic bacterial isolates against root knot nematode, Meloidogyne incognita in tomato under glasshouse condition. Int J Curr Microbiol Appl Sci 8(1):2584–2589. https://doi.org/10.20546/ijcmas.2019.801.271
Vurukonda SSKP, Giovanadri D, Stefani E (2021) Growth promotion and biocontrol activity of endophytic Streptomyces spp. prime archives in molecular sciences, 2nd Edition. pp.1–55. http://hdl.handle.net/11380/1248582
Węgrzyn A, Felis E (2018) Isolation of bacterial endophytes from Phalaris arundinacea and their potential in diclofenac and sulfamethoxazole degradation. Pol J Microbiol 67(3):321. https://doi.org/10.21307/pjm-2018-039
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173(2):697–703. https://doi.org/10.1128/jb.173.2.697-703.1991
Whipps JM (2001) Microbial interactions and biocontrol in the rhizosphere. J Exp Bot 52(1):487–511. https://doi.org/10.1093/jexbot/52.suppl_1.487
Wicaksono WA, Jones EE, Casonato S, Monk J, Ridgway HJ (2018) Biological control of Pseudomonas syringae pv. actinidiae (Psa), the causal agent of bacterial canker of kiwifruit, using endophytic bacteria recovered from a medicinal plant. Biol Control 116:103–112. https://doi.org/10.1016/j.biocontrol.2017.03.003
Wu T, Xu J, Liu J, Guo W-H, Li X-B, Xia J-B, Xie W-J, Yao Z-G, Zhang Y-M, Wang R-Q (2019) Characterization and initial application of endophytic Bacillus safensis strain ZY16 for improving phytoremediation of oil-contaminated saline soils. Front Microbiol 10:991. https://doi.org/10.1016/j.tifs.2019.08.017
Xia Y, Li S, Liu X, Zhang C, Xu J, Chen Y (2019) Bacillus halotolerans strain LYSX1-induced systemic resistance against the root-knot nematode Meloidogyne javanica in tomato. Ann Microbiol 69(12):1227–1233. https://doi.org/10.1007/s13213-019-01504-4
Xiang N, Lawrence KS, Donald PA (2018) Biological control potential of plant growth-promoting rhizobacteria suppression of Meloidogyne incognita on cotton and Heterodera glycines on soybean: a review. J Phytopathology 166(7–8):449–458. https://doi.org/10.1111/jph.12712
Xie Y, Nian L, Zeng Y, Wang M, Yuan B, Cheng S, Cao C (2021) Dynamic variation of endogenous flora in kiwifruit and its association with ripening metabolism in response to ethylene micro-environment. Postharvest Biol Technol 182:111695. https://doi.org/10.1016/j.postharvbio.2021.111695
Zhang L, Li W, Tao Y, Zhao S, Yao L, Cai Y, Niu Q (2019) Overexpression of the key virulence factor 1, 3–1, 4-β-D-glucanase in the endophytic bacterium Bacillus halotolerans Y6 to improve Verticillium resistance in cotton. J Agric Food Chem 67(24):6828–6836. https://doi.org/10.1021/acs.jafc.9b00728
Zhang D, Xu H, Gao J, Portieles R, Du L, Gao X, Borroto Nordelo C, Borrás-Hidalgo O (2021) Endophytic Bacillus altitudinis strain uses different novelty molecular pathways to enhance plant growth. Front Microbiol 12:1613. https://doi.org/10.3389/fmicb.2021.692313
Acknowledgements
Not applicable.
Funding
The present study was supported financially by Guilan University, Guilan, Iran.
Author information
Authors and Affiliations
Contributions
SJ (Plant Pathology-Nematology), MG (Plant Pathology-Bacteriology), and MG (Horticultural Science) were the advisor and the supervisors of the thesis; SNB carried out all the experiments (Plant Pathology Student—PhD thesis). All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
The manuscript has not been published elsewhere.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Banihashemian, S.N., Jamali, S., Golmohammadi, M. et al. Isolation and identification of endophytic bacteria associated with kiwifruit and their biocontrol potential against Meloidogyne incognita. Egypt J Biol Pest Control 32, 111 (2022). https://doi.org/10.1186/s41938-022-00601-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41938-022-00601-y