Abstract
Background
The two-spotted spider mite, Tetranychus urticae Koch (Trombidiformes: Tetranychidae), is one of the most damaging mites in agriculture. Due to the concern for the intensive use of synthetic acaricides, entomopathogenic fungi represents a feasible alternative to T. urticae management. In the present study, 7 isolates of Metarhizium were characterized physiological and molecularly (based on the ITS1-5.8s-ITS2 rDNA) and evaluated for their acaricidal activity [mortality, mean and 90 lethal concentration (LC50: LC90) and mean and 90 lethal time (LT50: LT90)] against T. urticae under laboratory conditions.
Results
Sequencing of the ITS1-5.8s-ITS2 rDNA region indicated that the 7 isolates belong to M. anisopliae. The isolates Ma114 (3.7 ± 0.006 mm day−1), Ma109 (3.5 ± 0.009 mm day−1) and Ma106 (3.5 ± 0.006 mm day−1) had the highest radial growth rate and Ma114 (92.2 ± 0.86%) and Ma108 (94.4 ± 1.07%) had the highest germination percentage. All isolates were pathogenic to T. urticae, causing mortality that ranged from 45.3 to 85.3%. The LC50 and LC90 were 1.2 and 2.8, 1.1 and 2.5, and 1.2 and 2.8 × 108 conidia mL−1 for isolates Ma110, Ma109 and Ma106, respectively, while the LT50 and LT90 were 7.7 and 16.5, and 7.2 and 16.1 days for isolates M110 and Ma109, respectively.
Conclusion
The isolates Ma110 and Ma109 of M. anisopliae were moderately pathogenic and virulent against T. urticae.
Similar content being viewed by others
Background
The two-spotted spider mite (Tetranychus urticae Koch) is one of the most damaging pests in agriculture. Damage by T. urticae is typical in leaves, but at high population density inflorescences and fruits are also affected. Infested plants undergo a decrease in photosynthetic rate, plant growth and fruit production (Landeros et al. 2013). To control T. urticae, synthetic acaricides have been intensively used, but even though this strategy has alleviated the problem to some extent, issues associated with the continuous use of these chemicals include a decrease in population of beneficial arthropods, increased risk for human health and selection of acaricide resistant populations (Shin et al. 2017).
The use of biological control agents such as entomopathogenic fungi (EPF) represents an alternative to reduce the dependence of synthetic acaricides to control of phytophagous mites. Among EPF, Metarhizium spp. (Ascomycota: Clavicipitaceae) are considered an effective biological control agent against a wide range of phytophagous mites (Souza et al. 2014). In laboratory assays, Metarhizium anisopliae has shown high activity against T. urticae with mortalities range from 80 to 100%, when mites were exposed to 1.0 × 107 to 1.0 × 108 conidia mL−1 (Dogan et al. 2017). In the field and greenhouse conditions, M. anisopliae at concentration of 1 × 108 conidia mL−1 had also showed efficacy to reduce the population of T. urticae (Bugeme et al. 2014a). Also, Metarhizium has been proven to cause mortality on insects from different orders including: Hemiptera, Heteroptera, Coleoptera, Lepidoptera, Thysanoptera, Orthoptera, Diptera and Isoptera (Carolino et al. 2014). In addition, it has already used in commercial formulation of bioinsecticides for pest insects of foliage, root and stored grain (Kepler and Rehner 2013). Due to the importance of the biological control of two-spotted spider mite, the objective of this study was to characterize physiological and molecularly fungal isolates of Metarhizium and evaluate their acaricidal activity against the T. urticae.
Methods
Fungal isolates
Seven isolates of Metarhizium were obtained from the Laboratory of Biological Control of the Faculty of Biological and Agricultural Sciences at the University of Colima in Mexico. The isolates were obtained from soil samples in the states of Colima and Jalisco, Mexico (Table 1). The fungi were grown on Sabouraud dextrose agar (SDA, MCD® Lab, Mexico) in Petri dishes under laboratory conditions 25 ± 1 °C. To obtain the spore suspension, the conidia were harvested from the surface of a fungal colony (12 days old) by scrapping using 10 mL of sterile distilled water plus Tween 80 (0.05%), and then, it was filtered with a sterile gauze to avoid the mycelium and to recover only the conidia (Chan-Cupul et al. 2010). The conidia suspension was used for the bioassays.
Molecular identification of Metarhizium spp.
Genomic DNA extraction was performed from monosporic cultures in GPY culture medium, according to the method developed in the GeMBio laboratory (Tapia-Tussell et al. 2006). The ITS1-5.8s-ITS2 region of the rDNA was amplified using the primers ITS1 (5′ TCCGTAGGTGAACCTGCGG 3′) and ITS4 (5′TCCTCCGCTTATTGATATGC 3′) (White et al. 1990), at a final volume of 50 µL, which they contained 25 ng of genomic DNA, 0.20 mM of each dNTP (Invitrogen), 1.5 mM of MgCl2, 1 µM of primers and 1 U of Taq DNA polymerase (Invitrogen). DNA amplification was performed in a GeneAmp 9700 DNA thermal cycler (Perkin-Elmer). PCR conditions were: 94 °C for 1 min; 30 cycles at 94 °C for 1 min, 58 °C for 1 min, and 72 °C for 1 min; and 5 min at 72 °C (Tapia-Tussell et al. 2008). The PCR products were sequenced at Macrogen Inc. (Seoul, Korea). The sequences were compared with the database of the gene bank of the National Center for Biotechnology Information (s. f.) in the Basic Local Alignment Search Tool (BLAST) program and a phylogenetic tree was elaborated with the MEGA software version 10 with the neighbor-joining (Kumar et al. 2018).
Physiological characterization
The radial growth and conidial germination were used as parameters of physiological characterization of the Metarhizium isolates (Permandi et al. 2020). To evaluate the radial growth rate (RGR), 5 μL of a conidial suspension of 1 × 106 conidia mL−1 of each isolate was inoculated individually in the center of a Petri dish with SDA. The conidia suspension was obtained from an 11-day-old colony in a Petri dish, 10 mL of sterile distilled water with Tween (0.05%) was deposited on the surface of the colony and scraped with a sterile spatula, and conidia were recovered by filtration in a falcon tube (50 mL) with sterile gauze. The conidia concentration was counted and adjusted in a Neubauer chamber. Petri dishes were incubated at 25 ± 1 °C and photoperiod of 16: 8 h light: darkness. The diameter of the growing colony was measured daily for 10 days using a Vernier, with the colony diameter values were calculate the radial growth rate (mm d−1) (Chan-Cupul et al. 2010). A Petri dish served as a replicate and 10 replicates per fungal isolate were used.
The evaluation of conidial germination of each isolate was evaluated using the same Petri dishes from the RGR test. Ten microliters of 1 × 106 conidia mL−1 were deposited on SDA Petri dishes. The plates were incubated at 25 °C. Conidia germination was recorded each 12 h by 3 times (36 h) in a microscope (40×), 100 conidia for each plate were counted. Germinated conidia were considered when the germ tube has the same length than the conidia (Ayala-Zermeño et al. 2015). Ten replicates (Petri dishes) were used per fungal isolates. All bioassays were repeated twice.
Acaricidal activity of fungal isolates
A stock culture of T. urticae was established on common bean plants (Phaseolus vulgaris L.) into an entomological cage under greenhouse conditions (minimum temperature) = 24.0 °C, maximum temp = 38.5 °C, average temp = 31.4 °C, minimum relative humidity (RH) = 43.15%, maximum RH = 73.97%, average RH = 58.5% and 12:12 h of light: darkness photoperiod). For bioassays, fungal spore suspensions were obtained by 15-day-old colonies grown in SDA as previously described by Ayala-Zermeño et al. (2015). To test the pathogenicity of the fungal isolates, an initial assessment was conducted using a spore concentration of 1 × 107 conidia mL−1. Then, for the most active fungal isolates, a range of spore concentrations (1 × 107, 1 × 106, 1 × 105 and 1 × 104 conidia mL−1) was evaluated as described by Jeyarani et al. (2011). Spore suspensions were sprayed until runoff onto both surfaces of P. vulgaris leaves using hand sprayer. The leaves were air-dried for 20 min under the laminar flow cabinet, and then placed on wet cotton wool in Petri dishes. Adult mites (20 individuals per Petri dish) were then placed onto the treated leaves. Control leaves were sprayed with sterile distilled water containing 0.05% Tween 80. Petri dishes were placed in an incubator at 25 ± 1 °C, 75 ± 5% RH, and photoperiod of 16:8 h light: darkness. Mortalities were recorded daily for 12 days. Dead mites were transferred and kept for 15 days in Petri dishes lined with moist filter paper to observe mycosis (Eken and Hayat 2009). Each Petri dish served as a replicate, 8 replicates per fungal isolate were used.
Data analysis
All experiments were set in a completed randomized design. Data subjected to analyses of variance were checked for normality and homoscedasticity. Significant differences (P < 0.05) among means were determined by the Tukey test. Mean and ninety lethal times (LT50 and LT90) and mean and 90 lethal concentrations (LC50 and LC90) were calculated using Probit analysis. All analyses were performed in the Statistical Package for Statgraphics®.
Results
Molecular identification
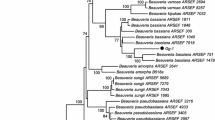
Sequencing of the ITS1-5.8s-ITS2 rDNA gene showed that the 7 isolates had homology with M. anisopliae (Metschn.) Sorokin 1883, 6 isolates with identity of 99.8–100% and the MA91 isolate of 99.2%, identity were obtained from BLAST algorithm NCBI database (Zhang et al. 2000). Phylogenetic analysis showed that isolates MA97, MA106, MA114 and MA109 were very close to each other, while isolates MA103 and MA110 shared the same clade, and isolate MA91 shared a clade with another isolated of M. anisopliae (Fig. 1).
Physiological characterization of fungal isolates
The radial growth (F = 83.01; df = 6; P < 0.001) and the germination of conidia (F = 120.69; df = 6; P < 0.001) varied significantly within fungal isolates. The highest radial growth rates were observed in the isolates Ma114 (3.7 mm day−1), Ma109 (3.5 mm day−1) and Ma106 (3.5 mm day−1). The germination of conidia was significantly higher in the isolate Ma114 (92%) relative to the other isolates, their values ranged from 62.6 to 84.6% (Table 2).
Acaricidal activity of fungal isolates
The mortality of T. urticae adults caused by Metarhizium isolates (1 × 107 conidia mL−1) varied significantly (F = 16.84; df = 7; P < 0.001). The highest mortalities of T. urticae were caused by the isolates Ma110 (83.5 ± 4.1%), Ma106 (80.8 ± 5.3%) and Ma109 (74.6 ± 7. 8%). In contrast, the isolates Ma103 (52.5%), Ma97 (48.3%) and Ma91 (56.4%) achieved the lowest mortalities (Fig. 2).
The mean lethal time (LT50) at the concentration of 1 × 107 conidia mL−1 for all Metarhizium isolates was calculated (Table 3). The lowest values of LT50 were achieved by the isolates Ma110 (7.7 days) and Ma109 (7.2 days); in contrast, the highest value of LT50 was achieved by the isolate Ma97 (13.2 days). The values of LT50 for the remaining isolates ranged from 9.7 to 11.7 days (Table 3). Regarding to the LT90, both the M. anisopliae isolates Ma110 and Ma109 achieved the lowest values with 16.5 and 16.1 days.
The isolates Ma110, Ma109 and Ma106 were evaluated in a dose–response experiment (1 × 104 to 1 × 107 conidia mL−1). Nonsignificant differences were observed among isolates for both, the LC50 and the LC90. The calculated LC50 for Ma110, Ma109 and Ma106 were 1.2, 1.1 and 1.2 × 108 conidia mL−1, respectively, and the calculated LC90 were 2.8, 2.5 and 2.8 × 108 conidia mL−1, for Ma110, Ma109 and Ma106, respectively (Table 4).
Discussion
In order to find a more safety method to control the two-spotted spider mite, this study evaluated the acaricidal activity of Metarhizium isolates against T. urticae. In addition, Metarhizium isolates were molecularly physiologically characterized. Phylogenetic analysis showed variability among strains, isolates Ma103 and Ma110 were grouped in the same clade, while isolate Ma91 was more distant and shared the clade with M. anisopliae KX756080. The distance among the isolates may suggest a new variety, as documented by Pantou et al. (2003) when amplified the ITS1-5.8S-ITS2 rDNA region of Metarhizium isolates. It must be taken into consideration that Metarhizium is a genus with a wide genetic diversity, because it is a cosmopolitan microorganism and adapts to different ecosystems and niches. In addition, it has different survival strategies, being a saprobic, entomopathogenic and endophyte genus (Ramírez-Milanes et al. 2022).
The physiological characterization of Metarhizium isolates indicated significant differences among isolates in radial growth and conidial germination. This result could be due by the genetic diversity of Metarhizium or at their biochemical activity, because fungal growth can be stimulated by the culture media (Gandarilla-Pacheco et al. 2012). In this regard, the present results are similar to those reported by Dimni et al. (2004) in M. anisopliae isolates, which achieved a mycelial growth from 2.3 to 3.2 mm day−1. In contrast, Nussenbaum et al. (2013) reported higher values for radial growth rate for Metarhizium isolates from 4.0 to 5.3 mm day−1.
Fungal growth rate is an important characteristic to select isolates for biological control programs, because it is expected that isolates with the highest growth rate would also have high acaricidal effects. Talaei-Hassanloui et al. (2007) suggested that there was a positive association between fungal radial growth and fungal virulence in EPF. Another important predictor of the acaricidal effect of a fungus is the conidial germination rate. In the present study, this variable ranged from 62.6 to 94.4% at 24 h post-inoculation. In another studies, Bugeme et al. (2009) observed in M. anisopliae 86–96% of conidial germination at 24 post-inoculation. Likewise, Onsongo et al. (2019) found similar results, and observed that the highest rate of conidial germination was achieved at temperatures of 25–30 °C. The rate of conidial germination is considered an important indicator of fungal virulence, based on the assumption that a spore on the insect cuticle that germinates rapidly would also have higher probability to penetrate and initiate the infection process in the host insect (Andersen et al. 2006); in M. anisopliae a positive relationship between fungal virulence and rate of conidial germination has been well documented (Ummidi et al. 2013). However, this asseveration is not a general rule, because in the present study M. anisopliae Ma114 achieved the highest RGR and conidia germination, but the isolate was not the most virulent.
All fungal isolates evaluated in this work caused significant mortality against T. urticae adults. Similar to other studies, the mortality caused by the fungal isolates ranged from 45.4 to 83.5% using 1 × 107 conidia mL−1. For example, Bugeme et al. (2014b) found 65 to 100% mortality using a spore concentration of 1 × 107 conidia mL−1. However, Chandler et al. (2005) observed that M. anisopliae isolates caused no more than 43% of mortality of T. urticae using 1 × 107 conidia mL−1.
In the present study, the calculated LT50 ranged from 7.2 to 13.2 days using a conidial suspension of 1 × 107 conidia mL−1. The lowest values for LT50 were observed for the isolates Ma110 (7.7 days) and Ma 109 (7.2 days). These values were relatively high compared to those reported in other studies, where calculated LT50 for Metarhizium spp. was within 2.2–4.0 days (Castro et al. 2018). Regarding to the LT90, Bugeme et al. (2009) reported values from 3.1 (ICIPE48) to 11.7 (ICIPE97) days for M. anisopliae isolates from Kenya. In other study, Hassan et al. (2017) reported 8.7 and 20.0 days for LT90 in M. anisopliae isolates from Egypt. In the tomato spider mite (Tetranychus evansi Baker & Pritchard), the LT90 values for M. anisopliae isolates ranged from 7.3 to 15.1 days (Vitalis et al. 2005), these values are less than those observed in the present study. These variations in LT50 and LT90 could be due to the genetic diversity of M. anisopliae; this genus is cosmopolitan with a versatile lifestyle as saprobe, endophyte, entomopathogen and antagonist of fungal plant pathogens. Recently, Serna-Domínguez et al. (2019) deported that Colima State has a wide genetic diversity of M. anisopliae including isolates with wide geographical distributions and different lifestyle.
By other hand, the calculated LC50 for the most pathogenic isolates (Ma110, Ma109 and Ma106) ranged from 1.1 × 108 to 1.2 × 108 conidia mL−1. These values are similar to those reported by Hassan et al. (2017), who documented LC50 of 9.3 × 107 and 4.57 × 108 conidia mL−1 for M. anisopliae isolates on T. urticae. In contrast, the LC50 values in the present study were relatively higher than the LC50 values of 2.0 × 105 to 5.0 × 105 conidia mL−1 reported in previous studies by Elhakim et al. (2020). Regarding the LC90, the isolates Ma110, Ma109 and Ma106 values that ranged from 2.5 to 2.8 × 108 conidia mL−1) were lower than those (LC90, 2.24 to 2.85 × 1010 conidia mL−1) reported by Elhakim et al. (2020). Taken both, the LT50 and LC50 as indicator of the fungal virulence, obtained fungal isolates may be considered moderately virulent. It is important to note that the virulence of EPF not only depends on intrinsic characteristics of the fungal isolates, but also on the concentration and frequency of the applications, as well as on the environmental conditions (Oyku et al. 2017).
Conclusions
Metarhizium anisopliae isolate Ma114 achieved the highest germination percentage and the isolates Ma114, Ma109 and Ma106 reached the highest mycelial growth rate. According to the sequencing of the ITS1-5.8s-ITS2 region (rDNA), the all studied isolates were identified as Metarhizium anisopliae (Metschn.) Sorokin 1883. The most pathogenic isolates were the M. anisopliae Ma110 and Ma106 that caused 83.46 and 80.76% mortality of T. urticae. According to the LT50, the most virulent M. anisopliae isolates against T. urticae were the Ma110 (7.7 days) and Ma109 (7.2 days). The calculated LC50 for the isolates Ma110 and Ma109 were 1.2 and 1.1 × 108 conidia mL−1, with nonsignificant difference between them.
Availability of data and materials
All data are available in the manuscript, and the materials used in this work are of high transparency and grade.
Abbreviations
- EPF:
-
Entomopathogenic fungi
- ITS:
-
Internal transcribed spacer
- rDNA:
-
Ribosomal deoxyribonucleic acid
- LC50 :
-
Mean lethal concentration
- LC90 :
-
Ninety lethal concentration
- LT50 :
-
Mean lethal time
- LT90 :
-
Ninety lethal time
- SDA:
-
Sabouraud dextrose agar
- Ma:
-
Metarhizium anisopliae
- PCR:
-
Polymerase chain reaction
- BLAST:
-
Basic local alignment search tool
- RGR:
-
Radial growth rate
- ANOVA:
-
Analyses of variance
- X2 :
-
Chi-square test
References
Andersen M, Magan N, Mead A, Chandler D (2006) Development of a population–based threshold model of conidial germination for analyzing the effects of physiological manipulation on the stress tolerance and infectivity of insect pathogenic fungi. Environ Microb 8:1625–1634. https://doi.org/10.1111/j.1462-2920.2006.01055.x
Ayala-Zermeño MA, Gallou A, Berlanga-Padilla AM, Serna-Domínguez MG, Arredondo-Bernal HC, Montesinos-Matías R (2015) Characterization of entomopathogenic fungi used in the biological control programme of Diaphorina citri in Mexico. Biocontrol Sci Technol 25:1192–1207. https://doi.org/10.1080/09583157.2015.1041878
Bugeme DM, Knapp M, Boga HI, Wanjova AK, Maniania NK (2009) Influence of temperature on virulence of fungal isolates of Metarhizium anisopliae and Beauveria bassiana to the two-spotted spider mite Tetranychus urticae. Mycopathologia 167:221–227. https://doi.org/10.1007/s11046-008-9164-6
Bugeme DM, Knapp M, Boga HI, Ekesi S, Maniania NK (2014a) Susceptibility of developmental stages of Tetranychus urticae (Acari: Tetranychidae) to infection by Beauveria bassiana and Metarhizium anisopliae (Hypocreales: Clavicipitaceae). Inter J Trop Insect Sci 34:190–196. https://doi.org/10.1017/S1742758414000381
Bugeme DM, Knapp M, Ekesi S, Chabi-Olaye A, Boga HI, Maniania K (2014b) Efficacy of Metarhizium anisopliae in controlling the two-spotted spider mite Tetranychus urticae on common bean in screenhouse and field experiments. Insect Sci 22:121–128. https://doi.org/10.1111/1744-7917.12111
Carolino AT, Paula AR, Silva CP, Butt TM, Samuels RI (2014) Monitoring persistence of the entomopathogenic fungus Metarhizium anisopliae under simulated field conditions with the aim of controlling adult Aedes aegypti (Diptera: Culicidae). Parasit Vectors 7:1–7. https://doi.org/10.1186/1756-3305-7-198
Castro T, Eilengerg J, Delalibera IJ (2018) Exploring virulence of new and less studied species of Metarhizium spp. from Brazil for two-spotted spider mite control. Exp Appl Acarol 74:139–146. https://doi.org/10.1007/s10493-018-0222-6
Chan-Cupul W, Ruiz-Sánchez E, Cristobal-Alejo J, Pérez-Gutiérrez A, Munguía-Rosales R, Lara-Reyna J (2010) In vitro development of four Paecilomyces fumosoroseus isolates and their pathogenicity on immature whitefly. Agrociencia 44:587–587
Chandler D, Davidson G, Jacobson RJ (2005) Laboratory and glasshouse evaluation of entomopathogenic fungi against the two-spotted spider mite, Tetranychus urticae (Acari: Tetranychidae), on tomato, Lycopersicon esculentum. Biocontrol Sci Technol 15:37–54. https://doi.org/10.1080/09583150410001720617
Dimni S, Maniania NK, Lux SA, Mueke JM (2004) Effect of constant temperatures on germination, radial growth and virulence of Metarhizium anisopliae to three species of African tephritid fruit flies. Biocontrol 49:83–94. https://doi.org/10.1023/B:BICO.0000009397.84153.79
Dogan YO, Hazir S, Yildiz A, Butt TM, Cakmak I (2017) Evaluation of entomopathogenic fungi for the control of Tetranychus urticae (Acari: Tetranychidae) and the effect of Metarhizium brunneum on the predatory mites (Acari: Phytoseiidae). Biol Control 111:6–12. https://doi.org/10.1016/j.biocontrol.2017.05.001
Eken C, Hayat R (2009) Preliminary evaluation of Cladosporium cladosporioides (Fresen.) de Vries in laboratory conditions, as a potential candidate for biocontrol of Tetranychus urticae Koch. World J Microbiol Biotechnol 25:489–492. https://doi.org/10.1007/s11274-008-9914-0
Elhakim E, Mohamed O, Elazouni I (2020) Virulence and proteolytic activity of entomopathogenic fungi against the twos potted spider mite, Tetranychus urticae Koch (Acari: Tetranychidae). Egypt J Biol Pest Control 30:1–8. https://doi.org/10.1186/s41938-020-00227-y
Gandarilla-Pacheco FL, Arévalo-Niño K, Galán-Wong LJ, Sandoval-Conrado CF, Quintero-Zapata I (2012) Evaluation of conidia production and mycelium growth in solid culture media from native strains of entomopathogenic fungi isolated from citrus-growing areas of Mexico. Afr J Biotechnol 11:14453–14460. https://doi.org/10.5897/AJB12.1658
Hassan DMA, Rizk MA, Sobhy HM, Mikhail WZA, Nada MS (2017) Virulent entomopathogenic fungi against the two-spotted spider mite Tetranychus urticae and some associated predator mites as non-target organisms. Egypt Acad J Biol Sci 10:37–56
Jeyarani S, Gulsar Banu J, Ramaraju K (2011) First record of natural occurrence of Cladosporium cladossporioides (Fresenius) de Vries and Beauveria bassiana (Bals.-Criv.) Vuill on two spotted spider mite, Tetranychus urticae Koch from India. J Entomol 8:274–279. https://doi.org/10.3923/je.2011.274.279
Kepler RM, Rehner SA (2013) Genome-assisted development of nuclear intergenic sequence markers for entomopathogenic fungi of the Metarhizium anisopliae species complex. Mol Ecol Resour 13:210–217. https://doi.org/10.1111/1755-0998.12058
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Landeros FJ, Cerna CE, Aguirre ULA, Flores CR, Ochoa FY (2013) Demographic parameters of Tetranychus urticae (Acari: Tetranychidae) on four Rosa sp. cultivars. Florida Entomol 96:1508–1512. https://doi.org/10.1653/024.096.0432
Nussenbaum AL, Lewylle MA, Lecuona RE (2013) Germination, radial growth and virulence to boll weevil of entomopathogenic fungi at different temperatures. World Appl Sci J 25:1134–1140. https://doi.org/10.5829/idosi.wasj.2013.25.08.1237
Onsongo SK, Gichimu BM, Akutse KS, Dubois T, Mohamed SA (2019) Performance of three isolates of Metarhizium anisopliae and their virulence against Zeugodacus cucurbitae under different temperature regimes, with global extrapolation of their efficiency. Insects 10:1–13. https://doi.org/10.3390/insects10090270
Oyku DY, Hazir S, Yildiz A, Butt TM, Cakmak I (2017) Evaluation of entomopathogenic fungi for the control of Tetranychus urticae (Acari: Tetranychidae) and the effect of Metarhizium brunneum on the predatory mites (Acari: Phytoseiidae). Biol Control 111:6–12. https://doi.org/10.1016/j.biocontrol.2017.05.001
Pantou MP, Mavridou A, Typas MA (2003) IGS Sequence variation, group-I introns and the complete nuclear ribosomal DNA of the entomopathogenic fungus Metarhizium: Excellent tools for isolate detection and phylogenetic analysis. Fungal Genet Biol 38:159–174. https://doi.org/10.1016/S1087-1845(02)00536-4
Permandi MA, Mukhlis B, Samosir BS, Siregar DY, Wayni M (2020) Physiology characterization of entomopathogenic fungi Beauveria bassiana and Metarhizium anisopliae on different carbohydrates sources. J Phys Conf Ser 1477:1–6
Ramírez-Milanes MN, Lezama-Gutiérrez R, Sánchez-Rangel JC, Chan-Cupul W, Buenrostro-Nava MT, Manzo-Sánchez G (2022) Genetic diversity of Metarhizium anisopliae isolated of insects and agroecosystems. Trop Subtrop Agroecosyst 25:1–8
Serna-Domínguez MG, Andrade-Michel GY, Rosas-Valdez R, Castro-Félix P, Arredondo-Bernal HC, Gallou A (2019) Genetic diversity of the Metarhizium anisopliae complex in Colima, Mexico, using microsatellites. Fungal Biol 123:855–863. https://doi.org/10.1016/j.funbio.2019.09.005
Shin TY, Bae SM, Kim DJ, Yun HG, Woo SD (2017) Evaluation of virulence, tolerance to environmental factors and antimicrobial activities of entomopathogenic fungi against two-spotted spider mite, Tetranychus urticae. Mycoscience 58:204–212. https://doi.org/10.1016/j.myc.2017.02.002
Souza R, Azevedo R, Lobo A, Rangel D (2014) Conidial water affinity is an important characteristic for termotolerance in entomopathogenic fungi. Biocontrol Sci Technol 24:448–461. https://doi.org/10.1080/09583157.2013.871223
Talaei-Hassanloui R, Kharazi-Pakdel A, Goettel MS, Little S, Mozaffari J (2007) Germination polarity of Beauveria bassiana conidia and its possible correlation with virulence. J Inverteb Pathol 94:102–107. https://doi.org/10.1016/j.jip.2006.09.009
Tapia-Tussell R, Lappe P, Ulloa M, Quijano-Ramayo A, Cáceres-Farfán M, Larqué-Saavedra A, Perez-Brito D (2006) A rapid and simple method for DNA extraction from yeasts and fungi isolated from Agave fourcroydes. Mol Biotechnol 33:67–70. https://doi.org/10.1385/MB:33:1:67
Tapia-Tussell R, Quijano-Ramayo A, Cortes-Velazquez A, Lappe P, Larque-Saavedra A, Pérez-Brito D (2008) PCR-based detection and characterization of the fungal pathogens Colletotrichum gloeosporioides and Colletotrichum capsici causing anthracnose in papaya (Carica papaya L.) in the Yucatan Peninsula. Mol Biotechnol 40:293–298. https://doi.org/10.1007/s12033-008-9093-0
Ummidi VRS, Josyula U, Vadlamani P (2013) Germination rates of Beauveria bassiana and Metarhizium anisopliae its possible correlation with virulence against Spodoptera litura larvae. Int J Adv Res 2:625–630
Vitalis WW, Nguya KM, Markus K, Hamadi IB (2005) Pathogenicity of Beauveria bassiana and Metarhizium anisopliae to the tobacco spider mite Tetranychus evansi. Exp Appl Acarol 36:41–50. https://doi.org/10.1007/s10493-005-0508-3
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sminsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press Inc, New York, pp 315–322. https://doi.org/10.1093/molbev/mst197
Zhang Z, Schwartz S, Wagner L, Miller W (2000) A greedy algorithm for aligning DNA sequences. J Comput Biol 7:203–214. https://doi.org/10.1089/10665270050081478
Acknowledgements
The first author wishes to thank the National Council of Science and Technology (CONACYT) for its doctoral fellowship.
Funding
No funding personal research.
Author information
Authors and Affiliations
Contributions
All authors contributed equally in the manuscript. MCB developed the bioassays and wrote the manuscript. ERS and WCC isolated and identified morphologically the fungal strains. TVY and RMM identified molecularly the fungal strains; ARR and HBG analyzed the collected data and wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable. This manuscript is in accordance with the guide for authors available on the journal’s website. Also, this work has not been published previously and is approved by all authors and host authorities. This article does not contain any studies with human participants or animals.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Cua Basulto, M., Ruiz Sánchez, E., Ballina Gómez, H. et al. Physiological and molecular characterization of Metarhizium isolates and their acaricidal activity against Tetranychus urticae Koch (Trombidiformes: Tetranychidae). Egypt J Biol Pest Control 32, 30 (2022). https://doi.org/10.1186/s41938-022-00530-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41938-022-00530-w