Abstract
Ensifer meliloti 4H41 is an aerobic, motile, Gram-negative, non-spore-forming rod that can exist as a soil saprophyte or as a legume microsymbiont of common bean (Phaseolus vulgaris). Strain 4H41 was isolated in 2002 from root nodules of P. vulgaris grown in South Tunisia from the oasis of Rjim-Maatoug. Strain 4H41 is salt- and drought-tolerant and highly effective at fixing nitrogen with P. vulgaris. Here we describe the features of E. meliloti 4H41, together with genome sequence information and its annotation. The 6,795,637 bp high-quality permanent draft genome is arranged into 47 scaffolds of 47 contigs containing 6,350 protein-coding genes and 72 RNA-only encoding genes, and is one of the rhizobial genomes sequenced as part of the DOE Joint Genome Institute 2010 Genomic Encyclopedia for Bacteria and Archaea-Root Nodule Bacteria (GEBA-RNB) project proposal.
Similar content being viewed by others
Introduction
Common bean ( Phaseolus vulgaris ) represents a very valuable source of proteins for low-income populations in Latin America and Africa [1]. However, this legume is considered to be a poor nitrogen-fixing pulse in comparison to other grain legumes [2]. This problem is generally attributed to the ineffectiveness of the native rhizobia, which is typically linked to the nodulation promiscuity of P. vulgaris [3-6] or to adverse abiotic conditions [7,8]. Salinity and drought are considered to be the major abiotic constraints that affect legumes in Tunisia and other countries. The selection of superior strains of rhizobia capable of assuring optimal nitrogen fixation under these adverse conditions is of high interest. Attention has therefore been directed to the isolation and characterization of rhizobial strains from various marginal areas that are subject to adverse climatic and edaphic conditions. In this context, Ensifer meliloti (formerly ‘ Sinorhizobium meliloti ’) strain 4H41 was isolated from root nodules of common bean grown in sandy, slightly alkaline soil from the oasis of Rjim-Maatoug in South Tunisia [9].
E. meliloti is classically considered to be a specific microsymbiont of the genera Medicago , Melilotus and Trigonella [10], however, recent studies have identified strains of E. meliloti that effectively nodulate P. vulgaris or several other legume species in northern Africa, South Africa and the Canary Islands [11-14]. Strain 4H41 induced nitrogen-fixing nodules on P. vulgaris but failed to nodulate Medicago spp. The phylogenetic analysis of nifH and nodC genes showed that strain 4H41 should be classified in a novel symbiovar (sv. mediterranense) [15]. The symbiovar mediterranense has also been used to describe Ensifer fredii and Ensifer americanum strains that can nodulate and fix nitrogen with P. vulgaris , species of Mexican Acacia (now reclassified as Vachellia and Senegalia [16]) and Leucaena leucocephala [17]. Strain 4H41 was able to grow in 4.4% NaCl (750 mM), while the P. vulgaris commercial inoculant Rhizobium tropici CIAT899T did not grow in salt concentrations higher than 1.8% [9]. Inoculationwater deficiency showed that strain 4H41 was more competitive and more effective than strain experiments under CIAT899T [7]. In field trials, P. vulgaris inoculated with strain 4H41 showed a significant increase in nodule number, shoot dry weight and grain yield even in non-irrigated fields. Under these conditions of water deficiency, nodulation by indigenous rhizobia was totally absent. However, when common bean was grown in adequately irrigated soil samples from these fields, numerous nodules could be observed, suggesting that, in contrast to 4H41, the native rhizobia were not tolerant of water deficiency [7]. Because of its effectiveness and high salt tolerance, strain 4H41 is considered to be an elite candidate for inoculant formulation in order to promote cultivation of common bean under salt and drought constraints. This strain has therefore been selected as part of the DOE Joint Genome Institute 2010 Genomic Encyclopedia for Bacteria and Archaea-Root Nodule Bacteria (GEBA-RNB) sequencing project [18]. Here we present a summary classification and a set of general features for E. meliloti strain 4H41, together with a description of its genome sequence and annotation.
Organism information
Classification and features
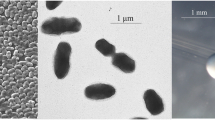
E. meliloti 4H41 is a motile, Gram-negative strain in the order Rhizobiales of the class Alphaproteobacteria . The rod shaped form (Figure 1 Left and Center) has dimensions of approximately 0.25-0.5 μm in width and 0.75-1.0 μm in length. It is fast growing, forming colonies within 3–4 days when grown on half strength Lupin Agar (½LA) [19], tryptone-yeast extract agar (TY) [20] or a modified yeast-mannitol agar (YMA) [21] at 28°C. Colonies on ½LA are white-opaque, slightly domed and moderately mucoid with smooth margins (Figure 1 Right).
Figure 2 shows the phylogenetic relationship of E. meliloti 4H41 in a 16S rRNA sequence based tree. This strain is the most similar to Ensifer meliloti LMG 6133T and Ensifer numidicus ORS 1407T based on the 16S rRNA gene alignment with sequence identities of 99.85% and 99.63%, respectively, as determined using the EzTaxon-e server [22]. Minimum Information about the Genome Sequence (MIGS) for 4H41 is provided in Table 1 and Additional file 1: Table S1.
Phylogenetic tree showing the relationship of Ensifer meliloti 4H41 (shown in bold blue print) to Ensifer spp. and other root nodule bacteria species in the order Rhizobiales, based on aligned sequences of the 16S rRNA gene (1,240 bp internal region). (The species name “Sinorhizobium chiapanecum” has not been validly published.) Azorhizobium caulinodans ORS 571T was used as an outgroup. All sites were informative and there were no gap-containing sites. Phylogenetic analyses were performed using MEGA, version 6 [45]. The tree was built using the Maximum-Likelihood method with the General Time Reversible model [46]. Bootstrap analysis [47] with 500 replicates was performed to assess the support of the clusters. Type strains are indicated with a superscript T. Strains with a genome sequencing project registered in GOLD [23] are in bold font and the GOLD ID is provided after the GenBank accession number, where this is available. Finished genomes are indicated with an asterisk.
Symbiotaxonomy
E. meliloti strain 4H41 is highly effective for nitrogen fixation with P. vulgaris , but is unable to nodulate several legume species that have previously been identified as E. meliloti hosts [14]. The symbiotic characteristics of E. meliloti strain 4H41 on a range of selected phylogenetically diverse hosts are provided in Table 2.
Genome sequencing information
Genome project history
This organism was selected for sequencing on the basis of its environmental and agricultural relevance to issues in global carbon cycling, alternative energy production, and biogeochemical importance, and is part of the Genomic Encyclopedia of Bacteria and Archaea, The Root Nodulating Bacteria chapter (GEBA-RNB) project at the U.S. Department of Energy, Joint Genome Institute (JGI). The genome project is deposited in the Genomes OnLine Database [23] and a high-quality permanent draft genome sequence is deposited in IMG [24]. Sequencing, finishing and annotation were performed by the JGI [25]. A summary of the project information is shown in Table 3.
Growth conditions and genomic DNA preparation
E. meliloti 4H41 was cultured to mid logarithmic phase in 60 ml of TY rich media [26] on a gyratory shaker at 28°C. DNA was isolated from the cells using a CTAB (Cetyl trimethyl ammonium bromide) bacterial genomic DNA isolation method [27].
Genome sequencing and assembly
The draft genome of E. meliloti 4H41 was generated at the DOE Joint Genome Institute (JGI) using the Illumina technology [28]. An Illumina standard shotgun library was constructed and sequenced using the Illumina HiSeq 2000 platform which generated 17,481,364 reads totaling 2,622.2 Mbp. All general aspects of library construction and sequencing performed at the JGI can be found on the JGI website [29]. All raw Illumina sequence data was passed through DUK, a filtering program developed at JGI, which removes known Illumina sequencing and library preparation artifacts [30]. The following steps were then performed for assembly: (1) filtered Illumina reads were assembled using Velvet (version 1.1.04) [31], (2) 1–3 Kbp simulated paired end reads were created from Velvet contigs using wgsim [32], (3) Illumina reads were assembled with simulated read pairs using Allpaths–LG (version r41043) [33]. Parameters for assembly steps were: 1) Velvet (velveth: 63 -shortPaired and velvetg: −very_clean yes –export-Filtered yes –min_contig_lgth 500 –scaffolding no –cov_cutoff 10) 2) wgsim (−e 0 –1 100 –2 100 –r 0 –R 0 –X 0) 3) Allpaths–LG (PrepareAllpathsInputs: PHRED_64 = 1 PLOIDY = 1 FRAG_COVERAGE = 125 JUMP_COVERAGE = 25 LONG_JUMP_COV = 50, RunAllpathsLG: THREADS = 8 RUN = std_shredpairs TARGETS = standard VAPI_WARN_ONLY = True OVERWRITE = True). The final draft assembly contained 47 contigs in 47 scaffolds. The total size of the genome is 6.8 Mbp and the final assembly is based on 830.5 Mbp of Illumina data, which provides an average 122.2x coverage of the genome.
Genome annotation
Genes were identified using Prodigal [34] as part of the DOE-JGI genome annotation pipeline [35,36]. The predicted CDSs were translated and used to search the National Center for Biotechnology Information (NCBI) nonredundant database, UniProt, TIGRFam, Pfam, KEGG, COG, and InterPro databases. The tRNAScanSE tool [37] was used to find tRNA genes, whereas ribosomal RNA genes were found by searches against models of the ribosomal RNA genes built from SILVA [38]. Other non–coding RNAs such as the RNA components of the protein secretion complex and the RNase P were identified by searching the genome for the corresponding Rfam profiles using INFERNAL [39]. Additional gene prediction analysis and manual functional annotation was performed within the Integrated Microbial Genomes Expert Review (IMG-ER) [40] developed by the Joint Genome Institute, Walnut Creek, CA, USA.
Genome properties
The genome is 6,795,637 nucleotides with 62.01% GC content (Table 4) and comprised of 47 scaffolds of 47 contigs. From a total of 6,422 genes, 6,350 were protein encoding and 72 RNA only encoding genes. The majority of protein-coding genes (82.01%) were assigned a putative function whilst the remaining genes were annotated as hypothetical. The distribution of genes into COGs functional categories is presented in Table 5.
Conclusion
Based on the 16S rRNA gene alignment, 4H41 is most closely related to Ensifer meliloti LMG 6133T , a Medicago microsymbiont [41] and Ensifer numidicus ORS 1407T , which effectively nodulates Argyrolobium uniflorum [42]. In contrast to these two strains, 4H41 is unable to nodulate either of these hosts. Strain 4H41 is one of 27 strains of E. meliloti with sequenced genomes deposited in the IMG database. Of these, 4H41 and strain GVPV12 [12] are the only two E. meliloti strains that have been isolated from, and are able to nodulate and fix nitrogen with, P. vulgaris . As the other sequenced E. meliloti strains are microsymbionts of Medicago spp., 4H41 is therefore a useful strain for comparing the molecular determinants of symbiosis in rhizobia with similar chromosomal backgrounds but which nodulate different legume hosts.
The genome size of the E. meliloti strains ranges from 6.6 – 8.9 Mbp; at 6.80 Mbp, the 4H41 genome is at the lower end of this range. It contains one pseudo gene, the numbers of which are highly variable in the sequenced E. meliloti genomes and can be up to 444 ( E. melilotiAK83). In common with the other E. meliloti genomes, 4H41 possesses a large number of genes assigned to COG functional categories for transport and metabolism of amino acids (12.22%), carbohydrates (11.03%), inorganic ions (5.3%), lipids (3.97%) and coenzymes (3.59%), and involved in transcription (8.78%), and signal transduction (3.58%). Genome analysis has revealed three distinct nodA genes, two coding for NodA proteins composed of 196 amino acids while the third encodes a NodA protein of 141 amino acids that lacks a 55 amino acid segment at the C-terminus. All three nodA copies are harboured within a symbiotic region of the genome and have highest sequence identity at the protein level with the common bean-nodulating strains Ensifer fredii GR64 [43] and E. meliloti GVPV12. Three distinct nodA genes are also found in the P. vulgaris commercial inoculant strains Rhizobium tropici CIAT 899 and Rhizobium sp. PRF 81 [44].
4H41 is salt- and drought-tolerant and highly effective for nitrogen fixation with P. vulgaris , and as such is a valuable inoculant strain. Analysis of its sequenced genome and comparison with the genomes of other sequenced E. meliloti and with RNB that nodulate the common bean will yield new insights into the molecular basis of salt- and drought-tolerance in rhizobia and into the molecular determinants of symbiotic specificity and nitrogen fixation in the important pulse legume P. vulgaris .
Abbreviations
- GEBA-RNB:
-
Genomic Encyclopedia for Bacteria and Archaea-Root Nodule Bacteria
References
Ribeiro RA, Barcellos FG, Thompson FL, Hungria M. Multilocus sequence analysis of Brazilian Rhizobium microsymbionts of common bean (Phaseolus vulgaris L.) reveals unexpected taxonomic diversity. Res Microbiol. 2009;160:297–306.
Hardarson G. Methods for enhancing symbiotic nitrogen fixation. Plant Soil. 1993;152:1–17.
Bromfield ESP, Barran LR. Promiscuous nodulation of Phaseolus vulgaris, Macroptilium atropurpureum, and Leucaena leucocephala by indigenous Rhizobium meliloti. Can J Microbiol. 1990;36:369–72.
Aouani ME, Mhamdi R, Mars M, Elayeb M, Ghtir R. Potential for inoculation of common bean by effective rhizobia in Tunisian soils. Agronomie. 1997;17:445–54.
Mhamdi R, Jebara M, Aouani ME, Ghrir R, Mars M. Genotypic diversity and symbiotic effectiveness of rhizobia isolated from root nodules of Phaseolus vulgaris L. grown in Tunisian soils. Biol Fertil Soils. 1999;28:313–20.
Thies JE, Singleton PW, Bohlool BB. Influence of the size of indigenous rhizobial populations on establishment and symbiotic performance of introduced rhizobia on field-grown legumes. Appl Environ Microbiol. 1991;57:19–28.
Mnasri B, Aouani ME, Mhamdi R. Nodulation and growth of common bean (Phaseolus vulgaris) under water deficiency. Soil Biol Biochem. 2007;39:1744–50.
Sessitsch A, Howieson JG, Perret X, Antoun H, Martínez-Romero E. Advances in Rhizobium research. Crit Rev Plant Sci. 2002;21:323–78.
Mnasri B, Mrabet M, Laguerre G, Aouani ME, Mhamdi R. Salt-tolerant rhizobia isolated from a Tunisian oasis that are highly effective for symbiotic N2-fixation with Phaseolus vulgaris constitute a novel biovar (bv. mediterranense) of Sinorhizobium meliloti. Arch Microbiol. 2007;187:79–85.
Fred EB, Baldwin IL, McCoy E. Root nodule bacteria and leguminous plants. Madison: University of Wisconsin Press; 1932.
Ardley JK, Reeve WG, O'Hara GW, Yates RJ, Dilworth MJ, Howieson JG. Nodule morphology, symbiotic specificity and association with unusual rhizobia are distinguishing features of the genus Listia within the southern African crotalarioid clade Lotononis s.l. Ann Bot. 2013;112:1–15.
Zurdo-Piñeiro JL, García-Fraile P, Rivas R, Peix A, León-Barrios M, Willems A, et al. Rhizobia from Lanzarote, the Canary Islands, that nodulate Phaseolus vulgaris have characteristics in common with Sinorhizobium meliloti isolates from mainland Spain. Appl Environ Microbiol. 2009;75:2354–9.
Fterich A, Mahdhi M, Caviedes M, Pajuelo E, Rivas R, Rodriguez-Llorente I, et al. Characterization of root-nodulating bacteria associated to Prosopis farcta growing in the arid regions of Tunisia. Arch Microbiol. 2011;193:385–97.
Mnasri B, Badri Y, Saidi S, de Lajudie P, Mhamdi R. Symbiotic diversity of Ensifer meliloti strains recovered from various legume species in Tunisia. Syst Appl Microbiol. 2009;32:583–92.
Rogel MA, Ormeño-Orrillo E, Martínez-Romero E. Symbiovars in rhizobia reflect bacterial adaptation to legumes. Syst Appl Microbiol. 2011;34:96–104.
Kyalangalilwa B, Boatwright JS, Daru BH, Maurin O, van der Bank M. Phylogenetic position and revised classification of Acacia s.l. (Fabaceae: Mimosoideae) in Africa, including new combinations in Vachellia and Senegalia. Bot J Linn Soc. 2013;172:500–23.
Mnasri B, Saïdi S, Chihaoui S-A, Mhamdi R. Sinorhizobium americanum symbiovar mediterranense is a predominant symbiont that nodulates and fixes nitrogen with common bean (Phaseolus vulgaris L.) in a Northern Tunisian field. Syst Appl Microbiol. 2012;35:263–9.
Reeve W, Ardley J, Tian R, Eshragi L, Yoon JW, Ngamwisetkun P, Seshadri R, Ivanova NN, Kyrpides NC. A genomic encyclopedia of the root nodule bacteria: Assessing genetic diversity through a systematic biogeographic survey. Stand Genomic Sci. 2015;10:14.
Howieson JG, Ewing MA, D'antuono MF. Selection for acid tolerance in Rhizobium meliloti. Plant Soil. 1988;105:179–88.
Beringer JE. R factor transfer in Rhizobium leguminosarum. J Gen Microbiol. 1974;84:188–98.
Vincent JM. A manual for the practical study of the root-nodule bacteria. International Biological Programme. Oxford, UK: Blackwell Scientific Publications; 1970.
Kim O-S, Cho Y-J, Lee K, Yoon S-H, Kim M, Na H, et al. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol. 2012;62:716–21.
Reddy TBK, Thomas AD, Stamatis D, Bertsch J, Isbandi M, Jansson J, et al. The Genomes OnLine Database (GOLD) v.5: a metadata management system based on a four level (meta)genome project classification. Nucleic Acids Res. 2014;43:D1099–106.
Markowitz VM, Chen I-MA, Palaniappan K, Chu K, Szeto E, Pillay M, et al. IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleic Acids Res. 2014;42:D560–7.
Mavromatis K, Land ML, Brettin TS, Quest DJ, Copeland A, Clum A, et al. The fast changing landscape of sequencing technologies and their impact on microbial genome assemblies and annotation. PLoS One. 2012;7:e48837.
Reeve WG, Tiwari RP, Worsley PS, Dilworth MJ, Glenn AR, Howieson JG. Constructs for insertional mutagenesis, transcriptional signal localization and gene regulation studies in root nodule and other bacteria. Microbiology. 1999;145:1307–16.
Joint Genome Institute Protocols. [http://jgi.doe.gov/collaborate-with-jgi/pmo-overview/protocols-sample-preparation-information/].
Bennett S. Solexa Ltd. Pharmacogenomics. 2004;5:433–8.
Joint Genome Institute Webpage. [http://jgi.doe.gov/].
Minkun L, Copeland A and Han J. DUK - A fast and efficient Kmer matching tool. [www.osti.gov/scitech/servlets/purl/1016000].
Zerbino DR, Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–9.
Github Wgsim. [https://github.com/lh3/wgsim].
Gnerre S, MacCallum I, Przybylski D, Ribeiro FJ, Burton JN, Walker BJ, et al. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc Natl Acad Sci U S A. 2011;108:1513–8.
Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119.
Mavromatis K, Ivanova NN, Chen IM, Szeto E, Markowitz VM, Kyrpides NC. The DOE-JGI Standard operating procedure for the annotations of microbial genomes. Stand Genomic Sci. 2009;1:63–7.
Chen IMA, Markowitz VM, Chu K, Anderson I, Mavromatis K, Kyrpides NC, et al. Improving microbial genome annotations in an integrated database context. PLoS One. 2013;8:e54859.
Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–64.
Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–96.
INFERNAL. Inference of RNA alignments [http://infernal.janelia.org/].
Markowitz VM, Mavromatis K, Ivanova NN, Chen IM, Chu K, Kyrpides NC. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics. 2009;25:2271–8.
De Lajudie P, Willems A, Pot B, Dewettinck D, Maestrojuan G, Neyra M, et al. Polyphasic taxonomy of rhizobia: Emendation of the genus Sinorhizobium and description of Sinorhizobium meliloti comb. nov., Sinorhizobium saheli sp. nov., and Sinorhizobium teranga sp. nov. Int J Syst Bacteriol. 1994;44:715–33.
Merabet C, Martens M, Mahdhi M, Zakhia F, Sy A, Le Roux C, et al. Multilocus sequence analysis of root nodule isolates from Lotus arabicus (Senegal), Lotus creticus, Argyrolobium uniflorum and Medicago sativa (Tunisia) and description of Ensifer numidicus sp. nov. and Ensifer garamanticus sp. nov. Int J Syst Evol Microbiol. 2010;60:664–74.
Torres Tejerizo G, Lozano L, González V, Bustos P, Romero D, Brom S. Draft genome sequence of the bean-nodulating Sinorhizobium fredii strain GR64. J Bacteriol. 2012;194:6978.
Ormeño-Orrillo E, Menna P, Almeida LG, Ollero FJ, Nicolás MF, Pains Rodrigues E, et al. Genomic basis of broad host range and environmental adaptability of Rhizobium tropici CIAT 899 and Rhizobium sp. PRF 81 which are used in inoculants for common bean (Phaseolus vulgaris L.). BMC Genomics. 2012;13:735.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol. 2011;28:2731–9.
Nei M, Kumar S. Molecular Evolution and Phylogenetics. New York: Oxford University Press; 2000.
Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–91.
Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, et al. Towards a richer description of our complete collection of genomes and metagenomes “Minimum Information about a Genome Sequence” (MIGS) specification. Nat Biotechnol. 2008;26:541–7.
Field D, Amaral-Zettler L, Cochrane G, Cole JR, Dawyndt P, Garrity GM, et al. The Genomic Standards Consortium. PLoS Biol. 2011;9:e1001088.
Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990;87:4576–9.
Editor L. Validation of publication of new names and new combinations previously effectively published outside the IJSEM. Int J Syst Evol Microbiol. 2005;55:2235–8.
Chen WX, Wang ET, Kuykendall LD. The Proteobacteria. New York: Springer - Verlag; 2005.
Garrity GM, Bell JA, Lilburn T. Class I. Alphaproteobacteria class. In: Garrity GM, Brenner DJ, Kreig NR, Staley JT, editors. Bergey's Manual of Systematic Bacteriology. 2nd ed. New York: Springer - Verlag; 2005.
Kuykendall LD. Order VI. Rhizobiales ord. nov. In: Garrity GM, Brenner DJ, Kreig NR, Staley JT, editors. Bergey’s Manual of Systematic Bacteriology. 2nd ed. New York: Springer - Verlag; 2005. p. 324.
Kuykendall LD. Family I. Rhizobiaceae. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT, editors. Bergey’s Manual of Systematic Bacteriology. New York: Springer - Verlag; 2005.
Kuykendall LD, Hashem FM, Wang ET. Genus VII. Sinorhizobium. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT, editors. Bergey’s Manual of Systematic Bacteriology. Volume 2. New York: Springer -Verlag; 2005. p. 358–61.
Judicial Commission of the International Committee on Systematics of Prokaryotes. The genus name Sinorhizobium Chen et al. 1988 is a later synonym of Ensifer Casida 1982 and is not conserved over the latter genus name, and the species name 'Sinorhizobium adhaerens' is not validly published. Opinion 84. Int J Syst Evol Microbiol. 2008;58:1973. doi:10.1099/ijs.0.2008/005991-0 [PubMed].
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9.
Guide to GO Evidence Codes | Gene Ontology Consortium. [http://geneontology.org/page/guide-go-evidence-codes].
GOLD Project ID for Ensifer meliloti 4H41. [https://gold.jgi-psf.org/project?id=10268].
Acknowledgements
This work was performed under the auspices of the US Department of Energy’s Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Berkeley National Laboratory under contract No. DE-AC02-05CH11231. We thank Gordon Thompson (Murdoch University) for the preparation of SEM and TEM photos.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contribution
RM supplied the strain, DNA and background information for this project, TR supplied DNA to JGI and performed all imaging, JA drafted the paper, RM provided financial support and all other authors were involved in sequencing the genome and/or editing the final paper. All authors read and approved the final manuscript.
Additional file
Additional file 1: Table S1.
Associated MIGS record.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Mhamdi, R., Ardley, J., Tian, R. et al. High-quality permanent draft genome sequence of Ensifer meliloti strain 4H41, an effective salt- and drought-tolerant microsymbiont of Phaseolus vulgaris . Stand in Genomic Sci 10, 34 (2015). https://doi.org/10.1186/s40793-015-0005-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40793-015-0005-1