Abstract
Background
Acquired primary chromosomal changes in cancer are sometimes found as sole karyotypic abnormalities. They are specifically associated with particular types of neoplasia, essential in establishing the neoplasm, and they often lead to the generation of chimeric genes of pathogenetic, diagnostic, and prognostic importance. Thus, the report of new primary cancer-specific chromosomal aberrations is not only of scientific but also potentially of clinical interest, as is the detection of their gene-level consequences.
Case presentation
RNA-sequencing was performed on a bone marrow sample from a patient with myelodysplastic syndrome (MDS). The karyotype was 46,XX,t(7;13)(p14;q12)[2]/46,XX[23]. The MDS later evolved into acute myeloid leukemia (AML) at which point the bone marrow cells also contained additional, secondary aberrations. The 7;13-translocation resulted in fusion of the gene PAN3 from 13q12 with PSMA2 from 7p14 to generate an out-of-frame PAN3–PSMA2 fusion transcript whose presence was verified by RT-PCR together with Sanger sequencing. Interphase fluorescence in situ hybridization analysis confirmed the existence of the chimeric gene.
Conclusions
The novel t(7;13)(p14;q12)/PAN3–PSMA2 in the neoplastic bone marrow cells could affect two key protein complex: (a) the PAN2/PAN3 complex (PAN3 rearrangement) which is responsible for deadenylation, the process of removing the poly(A) tail from RNA, and (b) the proteasome (PSMA2 rearrangement) which is responsible for degradation of intracellular proteins. The patient showed a favorable response to decitabine after treatment with 5-azacitidine and conventional intensive chemotherapy had failed. Whether this might represent a consistent feature of MDS/AML with this particular gene fusion, remains unknown.
Similar content being viewed by others
Background
Hematologic malignancies, including acute myeloid leukemia (AML) and acute lymphoblastic leukemia (ALL), often carry visible acquired chromosomal aberrations which may be either primary or secondary in leukemogenesis [1]. According to Heim and Mitelman [1] “Primary aberrations are frequently found as the sole karyotypic abnormalities in cancer and are often specifically associated with particular tumor types. The term primary not only refers to the fact that these are the first changes we see in neoplastic cells, but also reflects their causal role in tumorigenesis; they are essential in establishing the neoplasm”. In the same reference secondary aberrations are described as “rarely or never found alone; as the name implies, they develop in cells already carrying a primary abnormality. In later disease stages, however, they may be so numerous as to completely dominate the karyotypic picture. Although less specific than primary changes, secondary aberrations nevertheless demonstrate nonrandom features with distribution patterns that appear to be dependent both on which primary abnormality is present and on the type of neoplasm”.
The report of a new primary chromosomal aberration in a given cancer is of scientific as well as clinical interest. The aberration may give rise to a novel fusion protein, alternatively abrogation of an otherwise normal gene product, and thus define a new genetic subgroup in such malignancies [2]. An example is the cryptic t(7;21)(p22;q22)/RUNX1-USP42 chromosomal translocation which was first described in an AML patient and currently is considered a rare but nonrandom feature of myeloid malignancies where it is frequently found together with del(5q) [3,4,5,6,7].
We here present the molecular genetic and clinical features of a case of myelodysplastic syndrome with a novel primary t(7;13)(p14;q12) chromosome translocation that recombined the proteasome subunit alpha 2 (PSMA2) gene on 7p14 and the PAN3 poly(A) specific ribonuclease subunit (PAN3) gene on 13q12 generating a PAN3–PSMA2 fusion gene. The disease later evolved into AML at which point also secondary chromosome abnormalities could be seen.
Case presentation
A 74-year-old female patient was in April 2016 referred to our hospital because of thrombocytopenia. Blood analysis showed hemoglobin 10.8 g/dL, thrombocytes 31 × 109/L, and white blood cells 3.8 × 109/L with a normal differential count. Examination of a bone marrow biopsy showed 16% CD34-positive cells and dysplasia affecting mainly the megakaryocytic lineage, and the patient was diagnosed with myelodysplastic syndrome with excess of blasts 2 (MDS-EB 2). Cytogenetic analysis at this time showed the karyotype 46,XX,t(7;13)(p14;q12) (see below). No treatment was given.
One month later, examination of a new bone marrow aspirate showed 55% blasts and she was diagnosed with AML. Treatment with 5-azacytidine 5 days every 4th week was begun. However, a bone marrow aspirate after 6 monthly courses of 5-azacytidine showed 70% blasts, and because of disease progression she was now changed to intensive chemotherapy with age adjusted “3+7” (daunorubicin 50 mg/m2 day 1–3 and cytarabine 200 mg/m2 day 1–7), followed by one course of consolidation chemotherapy with mitoxantrone 10 mg/m2 day 1–5 and etoposide 100 mg/m2 day 1–5. In April 2017, 2 months after consolidation therapy, a new bone marrow aspirate again showed more than 50% blasts indicating AML recurrence. She was therefore started on decitabine 20 mg/m2 for 5 days every 4th week and after four courses, a new bone marrow biopsy showed 3–5% CD34+ cells. After another 2 months, a bone marrow biopsy still showed 5% CD34+ cells. Decitabine is continued and the patient is at the time of writing enjoying an active life in remission from her leukemia.
Bone marrow cells were cytogenetically investigated by standard methods [8, 9] and karyotyped according to the International System for Human Cytogenomic Nomenclature guidelines [10].
As part of our standard cytogenetic diagnosis of cases where AML is suspected, initial interphase FISH analyses of bone marrow cells were performed using the Cytocell multiprobe AML/MDS panel (Cytocell, http://www.cytocell.com) looking for -5/del(5q), -7/del(7q), del(20q), deletion of 17p13 (TP53), MLL rearrangements, PML-RARA fusion created by t(15;17)(q24;q11), RUNX1-RUNX1T1 created by t(8;21)(q22;q22), and CBFB-MYH11 generated by the inversion inv(16)(p13q22).
Further FISH analyses were performed using PAN3 and PSMA2 home-made break apart/double fusion probes. The BAC clones were purchased from BACPAC Resources Center (https://bacpacresources.org/home.htm). For the PAN3 gene on chromosome 13, the BAC clones were RP11-179F17 (accession number AL356915, position: chr13:28039868–28234287; band: 13q12.2; GRCh38/hg38 Assembly) and RP11-502P18 (accession number AL138712, position: chr13:28234288–28396806; band: 13q12.2–13q12.3; GRCh38/hg38 Assembly); they were labelled green. For the PSMA2 gene on chromosome 7, the BAC clones were RP11-111K18 (accession number ac010132, position: chr7:42801009–42967488; band: 7p14.1) and RP11-1081H3 (position: chr7:42923006–43086522; band: 7p14.1); they were labelled red. BAC DNA was extracted and probes were labelled with Fluorescein-12-dCTP (PerkinElmer, Boston, MA, USA) and Texas Red-5-dCTP (PerkinElmer) in order to obtain green and red signals, respectively, using Abbott’s nick translation kit (Des Plaines, IL, USA) and hybridized according to the companyʼs recommendations (http://www.abbottmolecular.com/home.html). FISH mapping of the clones on normal controls was performed to confirm their chromosomal location. Fluorescent signals were captured and analyzed using the CytoVision system (Leica Biosystems, Newcastle, UK).
Total RNA was extracted from the patientʼs bone marrow at the time when MSD was diagnosed using miRNeasy Mini Kit (Qiagen Nordic, Oslo, Norway). The RNA quality was evaluated using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) and 1 µg of total RNA was sent to the Genomics Core Facility at the Norwegian Radium Hospital, Oslo University Hospital (http://genomics.no/oslo/) for high-throughput paired-end RNA-sequencing. The RNA sequencing data were analyzed using the FusionCatcher software in order to discover fusion transcripts [11, 12] (https://github.com/ndaniel/fusioncatcher).
The primers used for PCR amplification and sequencing are listed in Table 1. The procedures of reverse transcriptase-polymerase chain reaction (RT-PCR) and direct sequencing of the PCR products were previously described [13]. For amplification of the PAN3–PSMA2 fusion transcript, the primer sets PAN3-957F1/PSMA2-206R1 and PAN3-1005F1/PSMA2-168R1 were used. For amplification of the putative reciprocal PSMA2–PAN3 fusion transcript, the primer set PSMA2-21F1/PAN3-1241R1 was used. For amplification of the PAN3 and PSMA2 transcripts, the primer sets PAN3-957F1/PAN3-1241R1 and PSMA2-21F1/PSMA2-206R1 were used, respectively. The PCR cycling involved initial denaturation at 94 °C for 30 s, followed by 35 cycles of 7 s at 98 °C, 30 s at 55 °C, 30 s at 72 °C, and a final extension for 5 min at 72 °C.
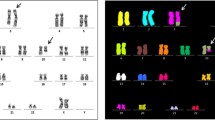
G-banding analysis of the bone marrow cells at diagnosis of MDS-EB2 yielded a karyotype with a single clonal abnormality: 46,XX,t(7;13)(p14;q12)[2]/46,XX[23] (Fig. 1a). G-banding analysis of a short-term cultured bone marrow aspirate drawn 4 months afterwards showed two cytogenetically related clones. In the first, two cells carried the chromosomal translocation t(7;13)(p14;q12) together with a der(13)ins(13;?)(q12;?). In the second subclone, ten cells carried a dic(19;20)(p13;q13) together with the two above-mentioned aberrations. The resulting karyotype was 46,XX,t(7;13)(p14;q12),der(13)ins(13;?)(q12;?)[2]/45,idem,dic(19;20)(p13;q13)[10]. G-banding analysis of a new bone marrow sample 9 months after the diagnosis yielded the karyotype 45,XX,t(7;13)(p14;q12),der(13)ins(13;?)(q12;?),dic(19;20)(p13;q13)[2]/46,XX[8]. Thus, G-banding analysis of bone marrow cells aspirated on several occasions indicated that t(7;13)(p14;q12) was the primary cytogenetic abnormality.
G-banding, molecular, and FISH analyses. a Partial karyotype showing the der(7) t(7;13)(p14;q12) and der(13)t(7;13)(p14;q12) together with the corresponding normal chromosome homologs. Breakpoint positions are indicated by arrows. b Gel electrophoresis showing the amplified cDNA fragments. c Partial sequence chromatogram of the amplified cDNA fragment showing the junction point of exon 6 of the PAN3 gene with exon 2 of PSMA2. d Deduced amino acid sequence of the PAN3–PSMA2 fusion transcript. The out-of-frame sequence from PSMA2 is in bold. The asterisk indicates the TAG termination codon. e Ideogram of chromosome 7 showing the mapping position of the PSMA2 gene (vertical red line). f Diagram showing the FISH probe for PSMA2. Additional genes in this region are also shown. g Ideogram of chromosome 13 showing the mapping position of the PAN3 gene (vertical green line). h Diagram showing the FISH probe for PAN3. Additional genes in this region are also shown. i FISH on interphase nucleus showing a red PSMA2 signal, a green PAN3 signal, and two yellow fusion signals
FISH analysis with multiprobes of the AML/MDS panel did not show -5/del(5q), -7/del(7q), del(20q), deletion of TP53, MLL rearrangements, or PML-RARA, RUNX1-RUNX1T1 or CBFB-MYH11 fusion (data not shown).
Using the FusionCatcher software on the fastq files of RNA sequencing data, 24 fusion genes were found (data not shown), among them fusion of PAN3 from chromosome band 13q12 with the PSMA2 gene from 7p14.
PCR with the primer combinations PAN3-957F1/PSMA2-206R1 and PAN3-1005F1/PSMA2-168R1 amplified 337 and 250 bp long cDNA fragments, respectively (Fig. 1b). Direct sequencing of the PCR products verified the presence of PAN3–PSMA2 (Fig. 1c). The fusion point was identical to that found by analysis of RNA sequencing data using FusionCatcher software (Fig. 1c). In the PAN3–PSMA2 transcript, exon 6 of PAN3 (nt 1152 in sequence with accession number NM_175854 version 7) was fused out-of-frame to exon 2 of PSMA2 (nt 90 in NM_002787 version 4) (Fig. 1c, d).
No cDNA amplified product was obtained when PSMA2 forward and PAN3 reverse primers (primer set PSMA2-21F1/PAN3-1241R1) were used, suggesting that the PSMA2–PAN3 fusion transcript was absent or not expressed (Fig. 1b). Both normal PAN3 (primer set PAN3-957F1/PAN3-1241R1) and PSMA2 (primer set PSMA2-21F1/PSMA2-206R1) cDNA fragments were amplified from the cDNAs of the patient suggesting that both genes are expressed in the bone marrow (Fig. 1b).
Using break apart/double fusion homemade probes for the PAN3 and PSMA2 genes (Fig. 1e–i), interphase FISH analysis was performed on cells from the second bone marrow sample with the karyotype 46,XX,t(7;13)(p14;q12),der(13)ins(13;?)(q12;?)[2]/45,idem,dic(19;20)(p13;q13)[10].
A green (PAN3 probe), a red (PSMA2 probe), and two yellow fusion signals were seen in 48 out of 68 examined nuclei (Fig. 1i).
Discussion and conclusions
We present here a case of MDS-EB2 progressing to AML in which a novel t(7;13)(p14;q12) was the primary acquired chromosomal change. At diagnosis of MDS, t(7;13) was the sole anomaly whereas 4 months later, when the patient had AML, the leukemic cells had acquired in their karyotype, in addition to t(7;13), the secondary changes der(13)ins(13;?)(q12;?) and dic(19;20)(p13;q13). RNA sequencing, RT-PCR, and FISH analyses showed that the t(7;13) rearranged the proteasome subunit alpha 2 (PSMA2) gene on 7p14 and the PAN3 poly(A) specific ribonuclease subunit (PAN3) gene on 13q12 to generate a PAN3–PSMA2 fusion gene.
PAN3 codes for the regulatory subunit of the poly(A)-nuclease (PAN) deadenylation complex (PAN2–PAN3 complex), one of two cytoplasmic eukaryotic poly(A) nuclease complexes involved in mRNA decay [14]. The PAN2–PAN3 complex specifically shortens poly(A) tails of RNA when the poly(A) stretch is bound by poly(A)-binding protein (PABP), followed by rapid degradation of the shortened mRNA tails by the CCR4-NOT complex [15, 16]. The N‐terminus of PAN3 contains a zinc finger and a PABP interacting motif 2 (PAM2). The C‐terminal part contains a pseudokinase, a coiled coil, and a C‐terminal knob domain [16,17,18,19,20,21]. The PAN3 pseudokinase domain binds ATP, a function required for mRNA degradation in vivo [16,17,18,19,20,21]. However, it does not have kinase activity due to structural rearrangements and loss of active site residues [16,17,18,19,20,21]. The coiled coil mediates dimerization and the C-terminal knob domain binds to the WD40 domain of PAN2 [16,17,18,19,20,21]. PAN3 acts as a positive regulator of PAN activity, recruiting the catalytic subunit PAN2 to mRNA via its interaction with PABP and to miRNA targets via its interaction with GW182 family proteins [16,17,18,19,20,21]. Heterozygous deletions of the PAN3 gene were reported in 5.4% of cases of highly hyperdiploid childhood acute lymphoblastic leukemia. Real-time quantitative RT-PCR analyses showed that in cases with deletion of PAN3, the gene was expressed at a lower level than in the samples without such deletion [22].
PSMA2 is ubiquitously expressed and encodes a peptidase which is a component of the alpha subunit of the 20S core proteasome complex [23]. The proteasome is a multi-catalytic proteinase complex. It is distributed throughout eukaryotic cells at a high concentration and cleaves peptides in an ATP/ubiquitin-dependent process as part of a non-lysosomal pathway [24, 25]. It plays a central role in the control of numerous cellular activities including regulation of the cell cycle [24, 25]. Inhibition of proteasome was found to be an effective therapeutic strategy in many hematologic malignancies [26,27,28,29].
The PAN3–PSMA2 fusion transcript codes for a putative PAN3 truncated protein which contains amino acid residues 1–333 from PAN3 protein (accession number NP_787050 version 6) corresponding to exons 1–3 of the gene, and 15 novel amino acid residues (ARLVNLSRLNMLWLL) stemming from the out-of-frame fusion of PSMA2. This putative protein would therefore contain the zinc finger and a PABP interacting motif 2 (PAM2) but would lack the normal C‐terminal part which contains the pseudokinase, the coiled coil, and the C‐terminal knob domain of PAN3. The precise role of the truncated PAN3 protein in the development of myelodysplasia/leukemia cannot be predicted without functional studies. However, an anomaly in deadenylation, which is fundamental to the regulation of gene expression, can be assumed. Alternatively, loss of a functional PAN3 and/or PSMA2 allele might be the important factor in pathogenesis. Whether any functional similarity exists between the present translocation and abrogation case and ALLs with deletion of an entire PAN3 allele [22], is a moot point.
Chromosomal rearrangements resulting in gene truncation have been described repeatedly for the RUNX1 and ETV6 genes [30, 31]. The aberrations generate a premature stop codon in the open reading frames leading to expression of C-terminal truncated forms of the RUNX1 or ETV6 proteins [30, 32]. Truncated RUNX1 proteins were shown to interfere with normal RUNX1 [33,34,35]. Truncated forms of ETV6 were found to have a dominant-negative effect on normal ETV6 function and disrupt both primitive and definitive hematopoiesis in the zebrafish model [36]. Chromosome translocations resulting in gene truncation have also been reported for other genes. For example, a t(3;21)(q22;q22) leading to truncation of RYK was seen in atypical chronic myeloid leukemia [37]. In a case of AML transformed from myelodysplastic syndrome, a t(2;7)(p24.3;p14.2) generated an out-of-frame NBAS-ELMO1 fusion transcript coding for a truncated NBAS protein [38]. Recently, in a case of AML, a t(3;5)(p24;q14) translocation was found to result in fusion of SATB1 with an expression sequence tag. The SATB1-fusion transcript would code for a SATB1 protein lacking the C-terminal DNA-binding homeodomain [39].
The patient in this report did not respond to treatment with 5-azacitidine, daunorubicin plus cytarabine, or mitoxantrone plus etoposide. She did, however, have a very favorable response to decitabine. It is possible that the 7;13-translocation is causatively involved in this difference, but in the absence of other cases with the same genetic change, one cannot tell. The case anyway illustrates that some patients with MDS or AML who do not respond favorably to standard treatment, may benefit from a change to decitabine [40, 41]. It may be particularly noteworthy from a clinical point of view that some patients who do not respond to 5-azacitidine, may do so to decitabine in spite of the fact that both drugs are hypomethylating agents.
Abbreviations
- ALL:
-
acute lymphoblastic leukemia
- AML:
-
acute myeloid leukemia
- BAC:
-
bacterial artificial chromosome
- FISH:
-
fluorescence in situ hybridization
- MDS:
-
myelodysplastic syndrome
- RT-PCR:
-
reverse transcriptase-polymerase chain reaction
References
Heim S, Mitelman F. Cancer cytogenetics: chromosomal and molecular genetic abberations of tumor cells. 4th ed. London: Wiley-Blackwell; 2015.
Panagopoulos I. From chromosomes to genes: searching for pathogenetic fusions in cancer. In: Heim S, Mitelman F, editors. Cancer cytogenetics: chromosomal and molecular genetic abberations of tumor cells. 4th ed. Chichester: Wiley-Blackwell; 2015. p. 42–61.
Foster N, Paulsson K, Sales M, Cunningham J, Groves M, O’Connor N, et al. Molecular characterisation of a recurrent, semi-cryptic RUNX1 translocation t(7;21) in myelodysplastic syndrome and acute myeloid leukaemia. Br J Haematol. 2010;148:938–43.
Jeandidier E, Gervais C, Radford-Weiss I, Zink E, Gangneux C, Eischen A, et al. A cytogenetic study of 397 consecutive acute myeloid leukemia cases identified three with a t(7;21) associated with 5q abnormalities and exhibiting similar clinical and biological features, suggesting a new, rare acute myeloid leukemia entity. Cancer Genet. 2012;205:365–72.
Ji J, Loo E, Pullarkat S, Yang L, Tirado CA. Acute myeloid leukemia with t(7;21)(p22;q22) and 5q deletion: a case report and literature review. Exp Hematol Oncol. 2014;3:8.
Panagopoulos I, Gorunova L, Brandal P, Garnes M, Tierens A, Heim S. Myeloid leukemia with t(7;21)(p22;q22) and 5q deletion. Oncol Rep. 2013;30:1549–52.
Paulsson K, Bekassy AN, Olofsson T, Mitelman F, Johansson B, Panagopoulos I. A novel and cytogenetically cryptic t(7;21)(p22;q22) in acute myeloid leukemia results in fusion of RUNX1 with the ubiquitin-specific protease gene USP42. Leukemia. 2006;20:224–9.
Czepulkowski B. Basic techniques for the preparation and analysis of chromosomes from bone marrow and leukaemic blood. In: Rooney DE, editor. Human cytogenetics: malignancy and acquired abnormalities. New York: Oxford University Press; 2001. p. 1–26.
Potter AM, Watmore A. Cytogenetics in myeloid leukaemia. In: Rooney DE, editor. Human cytogenetics: malignancy and acquired abnormalities. New York: Oxford University Press; 2001. p. 27–55.
McGowan-Jordan J, Simons A, Schmid M. ISCN 2016. an international system for human cytogenomic nomenclature basel. Karger. 2016.
Kangaspeska S, Hultsch S, Edgren H, Nicorici D, Murumagi A, Kallioniemi O. Reanalysis of RNA-sequencing data reveals several additional fusion genes with multiple isoforms. PLoS ONE. 2012;7:e48745.
Nicorici D, Satalan H, Edgren H, Kangaspeska S, Murumagi A, Kallioniemi O et al. FusionCatcher—a tool for finding somatic fusion genes in paired-end RNA-sequencing data. bioRxiv. 2014.
Panagopoulos I, Gorunova L, Zeller B, Tierens A, Heim S. Cryptic FUS-ERG fusion identified by RNA-sequencing in childhood acute myeloid leukemia. Oncol Rep. 2013;30:2587–92.
Brown CE, Tarun SZ Jr, Boeck R, Sachs AB. PAN3 encodes a subunit of the Pab1p-dependent poly(A) nuclease in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:5744–53.
Wahle E, Winkler GS. RNA decay machines: deadenylation by the Ccr4-not and Pan2–Pan3 complexes. Biochim Biophys Acta. 2013;1829:561–70.
Wolf J, Passmore LA. mRNA deadenylation by Pan2–Pan3. Biochem Soc Trans. 2014;42:184–7.
Christie M, Boland A, Huntzinger E, Weichenrieder O, Izaurralde E. Structure of the PAN3 pseudokinase reveals the basis for interactions with the PAN2 deadenylase and the GW182 proteins. Mol Cell. 2013;51:360–73.
Jonas S, Christie M, Peter D, Bhandari D, Loh B, Huntzinger E, et al. An asymmetric PAN3 dimer recruits a single PAN2 exonuclease to mediate mRNA deadenylation and decay. Nat Struct Mol Biol. 2014;21:599–608.
Schäfer IB, Rode M, Bonneau F, Schussler S, Conti E. The structure of the Pan2–Pan3 core complex reveals cross-talk between deadenylase and pseudokinase. Nat Struct Mol Biol. 2014;21:591–8.
Stubbs MT, Wahle E. Deadenylation-a piece of PANcake. EMBO J. 2014;33:1503–5.
Wolf J, Valkov E, Allen MD, Meineke B, Gordiyenko Y, McLaughlin SH, et al. Structural basis for Pan3 binding to Pan2 and its function in mRNA recruitment and deadenylation. EMBO J. 2014;33:1514–26.
Paulsson K, Forestier E, Lilljebjorn H, Heldrup J, Behrendtz M, Young BD, et al. Genetic landscape of high hyperdiploid childhood acute lymphoblastic leukemia. Proc Natl Acad Sci USA. 2010;107:21719–24.
Tamura T, Osaka F, Kawamura Y, Higuti T, Ishida N, Nothwang HG, et al. Isolation and characterization of alpha-type HC3 and beta-type HC5 subunit genes of human proteasomes. J Mol Biol. 1994;244:117–24.
Tanaka K. The proteasome: overview of structure and functions. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85:12–36.
Tomko RJ Jr, Hochstrasser M. Molecular architecture and assembly of the eukaryotic proteasome. Annu Rev Biochem. 2013;82:415–45.
Niewerth D, Dingjan I, Cloos J, Jansen G, Kaspers G. Proteasome inhibitors in acute leukemia. Expert Rev Anticancer Ther. 2013;13:327–37.
Tasian SK, Pollard JA, Aplenc R. Molecular therapeutic approaches for pediatric acute myeloid leukemia. Front Oncol. 2014;4:55.
Citrin R, Foster JB, Teachey DT. The role of proteasome inhibition in the treatment of malignant and non-malignant hematologic disorders. Expert Rev Hematol. 2016;9:873–89.
Csizmar CM, Kim DH, Sachs Z. The role of the proteasome in AML. Blood Cancer J. 2016;6:e503.
De Braekeleer E, Douet-Guilbert N, Morel F, Le Bris MJ, Basinko A, De Braekeleer M. ETV6 fusion genes in hematological malignancies: a review. Leuk Res. 2012;36:945–61.
Panagopoulos I, Torkildsen S, Gorunova L, Ulvmoen A, Tierens A, Zeller B, et al. RUNX1 truncation resulting from a cryptic and novel t(6;21)(q25;q22) chromosome translocation in acute myeloid leukemia: a case report. Oncol Rep. 2016;36:2481–8.
De Braekeleer E, Douet-Guilbert N, Morel F, Le Bris MJ, Ferec C, De Braekeleer M. RUNX1 translocations and fusion genes in malignant hemopathies. Future Oncol. 2011;7:77–91.
Hromas R, Busse T, Carroll A, Mack D, Shopnick R, Zhang DE, et al. Fusion AML1 transcript in a radiation-associated leukemia results in a truncated inhibitory AML1 protein. Blood. 2001;97:2168–70.
Ramsey H, Zhang DE, Richkind K, Burcoglu-O’Ral A, Hromas R. Fusion of AML1/Runx1 to copine VIII, a novel member of the copine family, in an aggressive acute myelogenous leukemia with t(12;21) translocation. Leukemia. 2003;17:1665–6.
Rodriguez-Perales S, Torres-Ruiz R, Suela J, Acquadro F, Martin MC, Yebra E, et al. Truncated RUNX1 protein generated by a novel t(1;21)(p32;q22) chromosomal translocation impairs the proliferation and differentiation of human hematopoietic progenitors. Oncogene. 2015;35:125–34.
Rasighaemi P, Liongue C, Onnebo SM, Ward AC. Functional analysis of truncated forms of ETV6. Br J Haematol. 2015;171:658–62.
Micci F, Panagopoulos I, Haugom L, Andersen HK, Tjonnfjord GE, Beiske K, et al. t(3;21)(q22;q22) leading to truncation of the RYK gene in atypical chronic myeloid leukemia. Cancer Lett. 2009;277:205–11.
Fujita K, Sanada M, Harada H, Mori H, Niikura H, Omine M, et al. Molecular cloning of t(2;7)(p24.3;p14.2), a novel chromosomal translocation in myelodysplastic syndrome-derived acute myeloid leukemia. J Hum Genet. 2009;54:355–9.
Torkildsen S, Brunetti M, Gorunova L, Spetalen S, Beiske K, Heim S, et al. Rearrangement of the chromatin organizer special AT-rich binding protein 1 gene, SATB1, resulting from a t(3;5)(p24;q14) chromosomal translocation in acute myeloid leukemia. Anticancer Res. 2017;37:693–8.
Lübbert M, Suciu S, Hagemeijer A, Rüter B, Platzbecker U, Giagounidis A, et al. Decitabine improves progression-free survival in older high-risk MDS patients with multiple autosomal monosomies: results of a subgroup analysis of the randomized phase III study 06011 of the EORTC Leukemia Cooperative Group and German MDS Study Group. Ann Hematol. 2016;95:191–9.
Welch JS, Petti AA, Miller CA, Fronick CC, O’Laughlin M, Fulton RS, et al. TP53 and decitabine in acute myeloid leukemia and myelodysplastic syndromes. N Engl J Med. 2016;375:2023–36.
Authorsʼ contributions
IP designed the research, performed experiments, and wrote the manuscript. LG interpreted the cytogenetics and FISH data. HKA performed cytogenetics and FISH experiments and interpreted the data. AB made the hematologic evaluations. AD made the hematologic evaluations. KA performed cytogenetics and FISH experiments and interpreted the data. FM interpreted the cytogenetics and FISH data. SH evaluated the cytogenetics and FISH data, and wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgements
The authors thank Nina Øino for excellent technical assistance.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All available data are included in the manuscript and its figures.
Consent for publication
Written informed consent was obtained from the patient to publication of the case details.
Ethics approval and consent to participate
The study was approved by the regional ethics committee (Regional komité for medisinsk forskningsetikk Sør-Øst, Norge, http://helseforskning.etikkom.no), and written informed consent was obtained from the patient to publication of the case details. The ethics committee’s approval included a review of the consent procedure. All patient information has been de-identified.
Funding
This work was supported by grants from the Norwegian Radium Hospital Foundation.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Panagopoulos, I., Gorunova, L., Andersen, H.K. et al. PAN3–PSMA2 fusion resulting from a novel t(7;13)(p14;q12) chromosome translocation in a myelodysplastic syndrome that evolved into acute myeloid leukemia. Exp Hematol Oncol 7, 7 (2018). https://doi.org/10.1186/s40164-018-0099-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40164-018-0099-4