Abstract
Background
Rickettsial diseases are considered important in public health due to their dispersal capacity determined by the particular characteristics of their reservoirs and/or vectors. Among the latter, fleas play an important role, since the vast majority of species parasitize wild and invasive rodents, so their detection is relevant to be able to monitor potential emerging diseases. The aim of this study was to detect, characterize, and compare Rickettsia spp. from the fleas of micromammals in areas with different human population densities in Chile.
Methods
The presence of Rickettsia spp. was evaluated by standard polymerase chain reaction (PCR) and sequencing in 1315 fleas collected from 1512 micromammals in 29 locations, with different human population densities in Chile. A generalized linear model (GLM) was used to identify the variables that may explain Rickettsia prevalence in fleas.
Results
DNA of Rickettsia spp. was identified in 13.2% (174 of 1315) of fleas tested. Fifteen flea species were found to be Rickettsia-positive. The prevalence of Rickettsia spp. was higher in winter, semi-arid region and natural areas, and the infection levels in fleas varied between species of flea. The prevalence of Rickettsia among flea species ranged between 0–35.1%. Areas of lower human density showed the highest prevalence of Rickettsia. The phylogenetic tree showed two well-differentiated clades with Rickettsia bellii positioned as basal in one clade. The second clade was subdivided into two subclades of species related to Rickettsia of the spotted fever group.
Conclusions
To our knowledge, this is the first report of the occurrence and molecular characterization of Rickettsia spp. in 15 flea species of micromammals in Chile. In this study, fleas were detected carrying Rickettsia DNA with zoonotic potential, mainly in villages and natural areas of Chile. Considering that there are differences in the prevalence of Rickettsia in fleas associated with different factors, more investigations are needed to further understand the ecology of Rickettsia in fleas and their implications for human health.

Similar content being viewed by others
Background
Rickettsia spp. are obligate intracellular microorganisms, Gram-negative coccobacilli, with the ability to reproduce, both in the nucleus and in the cytoplasm of infected cells [1]. These bacteria have a vertebrate reservoir and an arthropod vector (e.g. ticks, mites, fleas and lice); in some cases, the latter may be affected by these bacteria [2]. They have a worldwide distribution and are the causative agents of serious human infections [3].
Currently, 32 species are recognized (http://www.bacterio.net/-allnamesmr.html), and there are many strains that have not yet been characterized, while subspecies and uncultivated species are classified as “Candidatus” [4]. Recently, using new classification methods based on formal order analysis (FOA), which considers whole-genome sequencing analysis, two groups are recognized within the genus Rickettsia: the major typhus group (MTG) and major spotted fever group (MSFG). The MTG is divided into the typhus group (TG) and ancestral group (AG) and is transmitted by insects. MSFG includes the R. felis group, R. akari group, and the “classical” spotted fever group that includes several species transmitted by mites and hard ticks, of which the most important are R. rickettsii and R. conorii, that cause Rocky Mountain spotted fever and Mediterranean spotted fever, respectively [4]. Since Rickettsia research has focused on species that affect humans, other species have received less attention [5]. Thus, there are several species of rickettsiae identified and are exclusively associated with arthropods. They are without known secondary hosts and associated with other organisms such as herbivorous insects, leeches, amoebas, inclusive algae, and plants, indicating that these are more common than suspected [5, 6], and that the effects they could cause in humans when contact is made are unknown.
Worldwide, micromammals, and especially rodents, are the main flea hosts. It is recognized that 74% of known flea species parasitize them; therefore, rodents play a fundamental role in the spread of flea-borne diseases, as various species of rodent fleas can also parasitize humans [7]. In addition to this, many rodent species are capable of inhabiting wild environments and adapting to rural and urban environments, which could favor a continuous gradient of transmission between domestic and wild species, and humans [8, 9]. In Chile, despite the great diversity of described fleas (114 species), which mainly parasitize rodents [10, 11], a scarce number of studies have detected Rickettsia in fleas [12,13,14,15]. These studies have focused on the molecular detection of pathogens in fleas of domestic mammals, identifying R. felis from cat and dog fleas (Ctenocephalides felis and C. canis) in central (Metropolitan region) and southern Chile (Valdivia) [12,13,14]. Recently, “Candidatus Rickettsia asembonensis”, “Candidatus Rickettsia senegalensis”, and R. felis, were detected in C. felis from cats in the Easter Island (Rapa Nui) [15]. No studies have shown their presence in rodent fleas. If this adds to the expansion of the human population invading wild areas, the chance of contacting fleas on infected rodents increases. Since, in some places, peri-urban rodents provide a link between wild rodent and human communities, humans are exposed to some zoonotic agents that circulate in these natural ecosystems [16, 17].
The aim of this study was to detect, characterize, and compare Rickettsia spp. from the fleas of micromammals in areas with different human population densities in Chile. The findings will provide the baseline for the future surveillance of Rickettsia spp. in Chile.
Methods
Sample localities and micromammal-trapping procedures
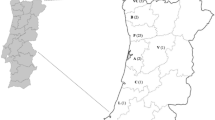
A total of 1512 micromammals belonging to 18 species (Table 1) were captured during a trapping effort of 11,034 trap/nights from 23 localities (9 cities, 6 villages and 8 natural areas) of the 29 sampled, covering 10 administrative regions in Chile and five bioclimatic regions (hyper-arid, arid, semi-arid, sub-humid and hyper-humid), latitude between −20.2167 and −53.1667 (Fig. 1). It was conducted from December 2015 to January 2018, during austral summer (December to February) and austral winter (July and September). These localities were selected based on the following demographic characteristics: (i) city, urban entity that has > 5000 inhabitants; (ii) village, urban entity with a population ranging between 2001–5000 inhabitants, or between 1001–2000 people, where less than 50% of the population that declares having worked, is engaged in primary activities (e.g. livestock, agriculture or fishing) [18]; and (iii) natural area, without human settlement, corresponding to national park (NP; unaltered areas of natural and biological diversity), and national reserves (NR; areas protecting wildlife populations or natural resources).
Micromammals were captured using a Sherman trap (23 × 7.5 × 9 cm, Sherman Co., Tallahassee, USA) and wire-mesh traps (30 × 10 × 11 cm; Forma Ltd., Santiago, Chile) baited with oats. The associated use of both types of traps strongly reduced the likelihood of a species being present but not captured. Each locality was sampled for two consecutive nights. In each sampling locality, the traps were placed in four parallel lines approximately 100 m from each other, and each line was equipped with 50 traps set 10 m apart from each other. Only in cities, traps were used along lines with a 5–10 m inter-trap space, and the traps were placed outside the buildings. The rodents were removed from the traps according to standard techniques [19], and were subsequently anesthetized with ketamine:xilazine (1:1) [20]. Flea samples from rodents were collected by hand or with forceps from the host and placed into sterile cryovials tubes with 95% ethanol. For each rodent, the total number of extracted fleas was recorded (abundance); with these data, the overall mean infection intensity (the number of fleas collected from all species/number of infested hosts), the overall mean abundance of infection (the number of collected fleas from all species/total number of hosts), and prevalence (the proportion of infected hosts) were calculated. The micromammals were identified following Iriarte [21]. Micromammals were released after sampling, except for invasive rodents [black rat (Rattus rattus), Norway rat (Rattus norvegicus), and house mouse (Mus musculus)] that were euthanized by cervical dislocation [19].
DNA extraction and PCR amplification
For DNA extraction, 5 fleas per host were selected, and when the number of fleas per host was less than 5, all the fleas were analyzed. Finally, DNA extraction was performed from 1315 fleas. Each flea was washed and cut between the third and fourth abdominal tergite with a scalpel. DNA was extracted from individual fleas using DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocols. The incubation time was 5 h; following DNA extraction, the flea’s exoskeleton was recovered and stored in 96% ethanol to later mount and identify the flea species.
The presence of Rickettsia spp. was initially screened by polymerase chain reaction (PCR) using a short fragment of citrate synthase (gltA) gene (401 bp; Table 2) [22]. Thereafter, gltA positive samples were tested using three genes: gltA (830 bp) [22], sca5 (ompB) [23], and we designed a set of primers for the β-subunit of RNA polymerase (rpoB) of Rickettsia sp. (GenBank: AF076436; Table 2). The amplification conditions were as follows: 5 min at 95 °C, 40 cycles of 30 s at 95 °C, 30 s of annealing temperature (see Table 2), 30 s at 72 °C, followed by a final extension of 5 min at 72 °C. The reactions were performed with GoTaq Green Master Mix 2X (Promega, Madison, USA) 12.5 µl, 5.5 µl of ultrapure nuclease-free water, 2 µl of forward primer (10 µM), 2 µl of reverse primer (10 µM), and 4 µl of DNA sample. The negative controls were carried out with ultrapure water, and the positive control was genomic DNA of R. conorii (AmpliRun® Rickettsia conorii DNA Control; Vircell, Granada, Spain). A selected number of Rickettsia-positive samples were purified and sequenced by the Macrogen Company (Seoul, Korea).
Phylogenetic and BLAST analyses
All DNA sequences were edited and aligned using the Codon Code Aligner (CodonCode Corporation, Centerville, MA, USA). All sequences generated in this study were compared with those available on GenBank using the BLAST program (see http://www.ncbi.nlm.nih.gov/BLAST/). A Bayesian probabilities tree was created using MrBayes 3.2 based on gltA 830-bp gene fragment, using Anaplasma phagocytophilum as the outgroup. We used the GTR + G substitution model to reconstruct the tree and 10,000,000 bootstrap trials.
Flea mounting and identification
After DNA extraction, each flea’s exoskeleton was recovered and mounted on glass slides using conventional procedures. The fleas were identified using a light microscope, taxonomic keys, and the descriptions of Johnson [24], and Sanchez & Lareschi [25]. Voucher specimens (slides) were catalogued in the Museo de Zoología at Universidad de Concepción (MZUC-UCCC, Concepción, Chile) under the accession numbers 46647–46667.
Statistical analysis
The prevalence (percentage of micromammals parasitized with fleas) and abundance mean (mean number of fleas per host) in species of micromammals was calculated with total of samples of fleas collected (n = 2272), and confidence intervals (95% CI) were calculated, using bootstrap (2000 bootstrap replicates). The prevalence of Rickettsia (percentage of fleas infected with Rickettsia) was calculated based on the PCR results. We used generalized linear models (GLM) with binomial distribution and logit function to identify the variables that may explain Rickettsia prevalence in fleas. The explanatory variables analyzed were bioclimatic regions (hyper-arid; arid; semi-arid; sub-humid; and hyper-humid), location type (city; village and natural area) and season (summer and winter). First, we built a model that included all bioclimatic regions and then we built models for each bioclimatic region. To assess the relationship between the prevalence and sample size, a Spearman correlation analysis was performed. The Chi-square test or Fisher’s exact test (if an expected cell count was < 5) was used to evaluate the differences in the prevalence of Rickettsia among species of flea. A P-value < 0.05 was considered statistically significant. The data were analyzed using JMP software® (SAS Institute Inc., Cary NC, USA).
Nucleotide sequence accession numbers
Rickettsia sequences generated in this study were deposited in the NCBI GenBank database under the following accession numbers: MN630893-MN630962 (gltA); MN630963-MN630997 (rpoB) and MT834938-MT834942 (sca5).
Results
A total of 2272 fleas were collected from 13 micromammal species, with an overall prevalence of 46.7% (n = 706). The overall mean abundance was 1.5 fleas per host and the overall mean intensity was 3.2 fleas per parasitized host (Table 1). Excluding the species in which < 20 individuals were sampled, the micromammals that presented the highest prevalence of fleas were Loxodontomys micropus (Austral greater mouse, 87.5%) and Octodon degus (Fence degu, 78.3%), and the lowest prevalence was found in R. rattus (29.2%). The abundance and mean intensity were higher in O. degus (Table 1). The marsupial Thylamys elegans (Llaca mouse-opossum) had a prevalence of fleas of 51.4%. All of the flea species found in T. elegans corresponded to species that were also found in rodents (Table 3).
Of all collected fleas, 1315 flea specimens were analyzed, corresponding to 27 species from 15 genera and 8 families (Table 4). The most abundant flea species were Sphinctopsylla ares (n = 211) and Neotyphloceras chilensis (n = 202; Table 4). The rodents that presented the greatest flea richness were Abrothrix olivacea (olive grass mouse, 17 spp.), R. rattus (14 spp.), A. hirta (long-haired grass mouse, 11 spp.), and Oligoryzomys longicaudatus (long-tailed pygmy rice rat, 11 spp.; Table 3). Natural areas were where the largest number of flea species (n = 25) and specimens were collected (n = 784), followed by villages (18 species, 349 specimens) and cities (18 species, 181 specimens). Agastopsylla boxi, Ctenoparia jordani, C. topali, Ectinorus cocyti and Plocopsylla lewisi were exclusive to natural areas (national parks and national reserves). Conversely, Xenopsylla cheopis was only found in one city (Iquique). Neotyphloceras chilensis and S. ares were the dominant species in natural areas (N. chilensis (n = 119); S. ares (n = 151)), and villages (N. chilensis (n = 83); S. ares (n = 50)), while Nosopsyllus fasciatus (n = 37), and C. inopinata (n = 25) were the most frequently collected in cities. Leptopsylla segnis, N. fasciatus, and X. cheopis are synanthropic rodent fleas [26], and were more abundant in cities than in villages and natural areas.
Rickettsiae prevalence on fleas
Fifteen flea species were found to be Rickettsia-positive for the short fragment (401 bp) of the gltA gene, 9 for the long fragment (830 bp) of the gltA gene, 10 for the rpoB gene, and 4 for the sca5 gene (Table 4). The highest prevalence (13.2%) was detected with the gltA 401-bp gene, followed by the rpoB (5.9%), gltA 830-bp (5.0%) and sca5 (0.5%) genes (Table 4). Among the flea species in which more than 20 individuals were analyzed, the prevalence varied between 0–35.1%. The Neotyphloceras spp. had the highest prevalence of Rickettsia (gltA 401-bp = 29.4%, gltA 830-bp = 9.56%, and rpoB = 11.25%; Table 4). The four fragments (gltA 401-bp, gltA 830-bp, rpoB and sca5) showed significant differences in the prevalence of detected Rickettsia (χ2 = 193.207, df = 3, P < 0.001), exception for gltA 830-bp and rpoB, which did not show significant differences (χ2 = 1.934, df = 1, P = 0.164). No association was found between the number of fleas analyzed and the prevalence of Rickettsia detected for any of the genes analyzed (rpoB: ρ = 0.4267, P = 0.12; gltA: ρ = 0.3757, P = 0.18; sca5: ρ = 0.3272, P = 0.35).
According to the GLM analysis, the prevalence of Rickettsia infection was significantly higher in the semi-arid region (27.8%). In addition, the overall prevalence was significantly higher in the winter (20.6%) than in the summer (5.3%). The prevalence of Rickettsia was higher in natural areas (15.9%), and cities exhibited a marginally significant lower prevalence (4.97%) compared to the other two location types (village: 11.2%; Table 5). Comparisons between bioclimatic regions showed that in the arid region, the prevalence of Rickettsia was higher in the natural areas and in the winter. While in the semi-arid region, the highest prevalence occurred in the winter (73.7%), and the highest prevalence of Rickettsia was detected in the natural areas (77.8%), differentiating from the cities (14.0%). In the sub-humid region, there was no effect of the factors on the prevalence of Rickettsia, whereas in the hyper-humid region, we detected Rickettsia (5.49%) only in the natural areas.
BLAST analysis and phylogenetic inference
A total of 167 sequences of gltA 401-bp (n = 68), gltA 830-bp (n = 40), rpoB (n = 54) and sca5 (n = 5) genes were analyzed (Table 6). For gltA 401-bp, out of the 68 sequences, 28 isolated from Delostichus phyllotis (n = 1), L. segnis (n = 1), N. crassispina (n = 1), N. pardinasi (n = 3), Neotyphloceras spp. (n = 7), N. fasciatus (n = 3), Plocopsylla sp. (n = 2), S. ares (n = 3), T. rhombus (n = 1) and Tetrapsyllus tantillus (n = 6) were 100% identical to Rickettsia sp. (GenBank: KY705378) obtained from the tick Amblyomma parvitarsum. Another 19 gltA sequences (401-bp) detected in Neotyphloceras spp. (n = 16), Chiliopsylla allophyla (n = 2) and C. inopinata (n = 1) were closely related to Rickettsia sp. MEAM1 (99%; GenBank: CP016305) isolated from whitefly Bemisia tabaci (Hemiptera: Aleyrodidae) (n = 16) and Rickettsia sp. Gr15 (GenBank: KP675966) detected in the tick Hyalomma marginatum (n = 3). Twenty-one sequences amplified from Neotyphloceras spp. (n = 1), S. ares (n = 13) and T. rhombus (n = 6) showed 97–98% identity with Rickettsia sp. (GenBank: U59712) isolated from Adelia bipunctata (Coleoptera: Coccinellidae). One sequence amplified from S. ares showed 93% similarity with uncultured Rickettsia sp. (GenBank: KY433588) detected in a tick.
Two sequences of gltA 830-bp segments showed high identity (99%) to “Candidatus Rickettsia senegalensis” (GenBank: KU499847) previously identified in a cat flea (C. felis). Forty sequences obtained from S. ares (n = 12), T. rhombus (n = 6), Neotyphloceras spp. (n = 19) and C. inopinata (n = 1) shared 97–98% identity with Rickettsia spp. (GenBank: KF646706; KY799066; U76908; and AJ269522) isolated from the insects Nesidiocoris tenuis (Heteroptera: Miridae), Mansonia uniformis (Diptera: Culicidae), Empoasca papayae (Hemiptera: Cicadellidae) and Adalia decempunctata (Coleoptera: Coccinellidae).
Seventeen amplified rpoB sequences in Neotyphloceras spp. shared 93–100% similarity with Rickettsia sp. MEAM1 (GenBank: CP016305) isolated from B. tabaci. Another 24 sequences derived from C. allophyla (n = 2), C. inopinata (n = 1), Neotyphloceras spp. (n = 1), S. ares (n = 14) and T. rhombus (n = 6) showed between 91% and 100% homology with Rickettsia sp. (GenBank: JF966777) of Synosternus pallidus (Siphonaptera: Pulicidae). Nine amplified sequences from Neotyphloceras spp. (n = 9) were 94–96% similar to Rickettsia sp. (GenBank: KX300157) isolated from a bat (Myotis emarginatus). Finally, 4 sequences isolated from Neotyphloceras spp. (n = 3) and T. rhombus (n = 1) showed lower homology with Rickettsia sp. (94%, GenBank: KX300203) isolated from a bat (Eptesicus serotinus).
Three sca5 fragments isolated from C. allophyla (n = 2) and C. inopinata (n = 1) showed homology with R. felis (94%; GenBank: GQ385243), and 2 fragments detected from S. ares showed low identity to R. hoogstraalii (GenBank: EF629536) (Table 6).
The phylogenetic tree shows two well-differentiated clades with 100% nodal support (Fig. 2). Clade R1 was formed by sequences obtained from Neotyphloceras fleas collected in Las Chinchillas NR (31°30′36″S, 71°05′15″W), Canela Baja (31°23′54″S, 71°27′27″W), and Fray Jorge NP (30°23′S, 71°23′W). Rickettsia bellii (GenBank: DQ146481) was positioned on a basal branch in this group. The clade R2 was subdivided into two subclades: R2a and R2b. R2a, with 93% nodal support, is related to sequences obtained from T. rhombus and S. ares collected in Los Queules NR, Cobquecura, and Coyhaique NR, comprising a larger area of distribution (latitude: − 35° to − 45°S) than clade R1. Subclade R2b was formed by sequences obtained from C. inopinata and C. allophyla collected in Los Queules NR and Nonguén NR, respectively. The newly generated sequences were positioned closely to R. hoogstraalii (GenBank: FJ767737) isolated from Haemaphysalis sulcata (tick) in Croatia [27], R. asembonensis detected in C. felis from Peru (GenBank: KY650697) [28] and R. felis isolated from C. felis in Brazil (GenBank: JN375498) [29].
Phylogenetic tree of gltA 830-bp gene of Rickettsia. The values on each node show the Bayesian probability of each clade. The accession number for each sequence is indicated. Flea species and locality are indicated for the sequences generated in this study. The principal clades are labelled R1, R2a and R2b
Discussion
To the best of our knowledge, we have provided for the first time evidence for the presence of Rickettsia DNA in 15 flea species identified on wild micromammals and synanthropic rodents in Chile. The prevalence of Rickettsia spp. infections in fleas varied between species of flea, bioclimatic regions, seasons and location type. We found a higher prevalence in winter, the semi-arid region and natural areas.
The fleas were characterized as being highly host-opportunistic, occupying various host species [7]. This is confirmed by our study, since of the 27 flea species collected, 19 parasitized more than one species of micromammal. We also highlight the high flea species richness recorded in R. rattus, where 10 of the 14 species identified in this rodent correspond to the flea species identified on native rodents. This rodent was mainly captured in urban areas; however, we also found it in rural and natural areas, this occurs mainly because these rodents have an omnivore diet and plasticity in their behavior, characteristics that allow them to inhabit a great diversity of environments, adapting successfully to urban, rural and wild environments [30, 31]. Rickettsia-positive fleas parasitizing R. rattus in these three areas indicate that this species could play a key role in spreading the disease from wild to urban environments [16, 32]. Conversely, we also observed that wild species enter human-occupied environments since they provide shelter and food. Abrothrix olivacea was the most frequently captured wild species in urban and rural areas and had the highest flea richness and the highest number of Rickettsia-positive fleas. This species has been described to have a “random walk” type of dispersal behavior, so it can easily go from wild to domestic environments [33]. These findings are important because these rodent species could act as “bridge hosts” and aid in the spread of the disease [32, 34]. On the other hand, in natural areas, the rodent species most frequently captured was A. hirta; this species, like A. olivacea, had a high prevalence of Rickettsia-positive fleas. This rodent decreased its presence in areas with human intervention, which is consistent with the findings reported by Monteverde & Hadora [33], who described that this rodent preferably moves within the wild environment. Rodent populations can act as “source populations” and may be involved in the direct transmission of the pathogen to the target population [34].
The prevalence of Rickettsia spp. infections detected in our study was variable (0–35%), and associated with the identity of the flea species, season, type of locality and bioclimatic area. However, similar differences have been reported in other studies. For example, Radzijevskaja et al. [35] reported different prevalence related to the flea species analyzed (range: 0–43%). Also, Kuo et al. [36] carried out an extensive sampling analyzing the presence of Rickettsia in six species of flea, reporting 0–12.1% of prevalence in the different species of flea analyzed. Furthermore, flea infestations in this study were generally higher during the winter; however, this did not occur in all bioclimatic areas. Other studies have found similar results, attributing this variation to the differences in the seasonal reproductive cycles of the different species of flea [37], which are unknown in most of the species found in this study. On the other hand, the higher prevalence of Rickettsia in fleas detected in natural areas can be explained by the greater diversity of species of micromammals and, therefore, of fleas. Thus, the differences in the prevalence of infection in the different species of flea, localities, seasons and bioclimatic zones found in our study, reveal the importance of the composition of the community, both fleas, and hosts, in determining the prevalence of Rickettsia in fleas, and therefore in the risk of infection in areas with different human disturbance.
In this study, we found two well-differentiated clades with a high degree of support. Clade R1 is formed by sequences obtained from fleas of the genus Neotyphloceras, collected from rodents Phyllotis darwini, A. olivacea, O. degus, R. rattus, and the marsupial T. elegans from central-north Chile (latitude: − 30° to − 31°S). This clade is related to R. bellii and is described as an ancestral group of Rickettsia [38], and which exhibits some specificity concerning its host [39]. This supports our results, where only bacteria detected in Neotyphloceras were found in this clade. Rickettsia bellii is endosymbiont of hard (Ixodidae) and soft (Argasidae) ticks throughout the American continent [39]. It has been classified as non-pathogenic for animals and humans [40], although seropositive samples have been found in dog blood in Brazil; however, the pathogenic effect is unknown [41]. Experimentally, this bacterium grows easily in mammalian cells. In experimental inoculations in guinea pig and rabbit, it produces, depending on the inoculated dose received, from a mild inflammatory reaction to necrotic scabs a typical symptomatology of other pathogenic rickettsiae [29]. Furthermore, it is capable of producing antibodies in experimental infections in the big-eared opossum Didelphis aurita, but without rickettsemia [42]. These results indicate that some flea species present in wild and synanthropic micromammals could carry a new ancestral genotype of Rickettsia, just like those reported by Song et al. [43] in China from fleas of wild rodents.
The R2 clade was divided into two large groups, R2a and R2b. R2a grouped all of the sequences detected in fleas being extracted from two species of flea, S. ares (Stephanocircidae) and T. rhombus (Rhopalopsyllidae), which were obtained from villages and natural environments through wide latitudinal distribution (latitude of − 35° to − 45°S). This corresponds to the wide distribution of the hosts of infected fleas (A. hirta and A. olivacea). Conversely, R2b was formed by sequences obtained from C. allophyla and C. inopinata belonging to the same family (Hystricopsylidae); both species of flea were collected in wild rodents (A. hirta and A. olivacea) from wild areas (Los Queules NR and Nonguén NR) in the south-central zone of Chile. These sequences are closely related to R. hoogstraalii, R. asembonensis and R. felis, all of which are members of the spotted fever group rickettsiae (SFG) [28, 29, 38]. The SFG consists of > 30 species that can be found worldwide, most of them with pathogenic effects on humans [44]. Our analysis showed a close relationship with R. hoogstraalii, a widely distributed bacterium that is still unknown for its pathogenicity in humans. This bacterium has been detected in both hard ticks (H. punctata, H. sulcate and H. parva) and soft ticks (Ornithodoros moubata, Carios capensis, C. sawaii and Argas persicus) present in domestic animals, bird nests, vegetation, and human dwellings [3, 45,46,47]. A similar situation occurs with R. asembonensis. It also has a wide distribution worldwide, having been reported in North America and South America, Asia, the Middle East and Europe [48], although it is associated with a greater number of ectoparasites, including fleas, ticks, and mites of domestic and peridomestic animals (C. canis, C. felis, X. cheopis, Pulex irritans, Amblyomma ovale, Rhipicephalus sanguineus, R. microplus and Ornithonysus bacoti) [49,50,51,52,53]. It has also been detected in monkey blood in Malaysia [54] and in dog blood in South Africa [55]. Although these bacteria live in parasitic arthropods close to humans and are closely associated with R. felis, there is no evidence yet of possible infection or pathogenicity [48]. On the other hand, R. felis is an emergent, widely distributed, flea-borne human pathogen, and like R. asembonensis and R. hoogstraalii, is associated with domestic and peridomestic animals and their ectoparasites [56, 57]. The main vector is C. felis, although mosquitoes (Anopheles gambiae) have also been detected as competent vectors [58]. Unlike R. asembonensis and R. hoogstraalii, this bacterium is of known pathogenicity causing fever, fatigue, nausea, muscle aches, back pain, headaches, macular rash, joint pain and eschar [49]. Although the BLAST analysis showed a low percentage of similarity with R. felis (sca5 94%), the phylogenetic analysis shows a close relationship with Rickettsia detected in C. allophyla in south-central Chile. Until now, in Chile, only R. felis has been registered in C. felis [12].
Conclusions
To the best of our knowledge, our study reports, for the first time in Chile, the presence of Rickettsia in different species of parasitic fleas of wild micromammals and invasive rodents found in both natural and human environments. Moreover, there is evidence of at least two clades of Rickettsia associated with fleas. These data increase the knowledge of possible Rickettsia vectors/reservoirs in Chile. However, greater efforts should be made to monitor and determine the degree of pathogenicity of the detected rickettsiae.
Availability of data and materials
Not applicable.
Abbreviations
- MTG:
-
major typhus group
- MSFG:
-
major spotted fever group
- TG:
-
typhus group
- AG:
-
ancestral group
- NP:
-
national park
- NR:
-
national reserve
- GLM:
-
generalized linear models
- SAG:
-
Servicio Agricola y Ganadero
- CONAF:
-
Corporación Nacional Forestal
- PCR:
-
polymerase chain reaction
- BLAST:
-
Basic Local Alignment Search Tool
- MZUC-UCCC:
-
Museo de Zoología Universidad de Concepción
- CI:
-
confidence interval
- n :
-
number of individuals
- df :
-
degrees of freedom
References
Walker DH. Rickettsia. In: Quah SR, editor. International encyclopedia of public health. 2nd ed. Amsterdam: Academic Press; 2016. p. 370–7.
Schumacher L, Snellgrove A, Levin ML. Effect of Rickettsia rickettsii (Rickettsiales: Rickettsiaceae) infection on the biological parameters and survival of its tick vector—Dermacentor variabilis (Acari: Ixodidae). J Med Entomol. 2016;53:172–6.
Blanton LS, Walker DH. Flea-borne rickettsioses and rickettsiae. Am J Trop Med Hyg. 2017;96:53–6.
Shpynov SN, Fournier PE, Pozdnichenko NN, Gumenuk AS, Skiba AA. New approaches in the systematics of rickettsiae. New Microbes New Infect. 2018;23:93–102.
Perlman SJ, Hunter MS, Zchori-Fein E. The emerging diversity of Rickettsia. Proc R Soc. 2006;273:2097–106.
Weinert LA, Werren JH, Aebi A, Stone GN, Jiggins FM. Evolution and diversity of Rickettsia bacteria. BMC Biol. 2009;7:6.
Bitam I, Dittmar K, Parola P, Whiting MF, Raoult D. Fleas and flea-borne diseases. Int J Infect Dis. 2010;14:e667–76.
Feng AYT, Himsworth CG. The secret life of the city rat: a review of the ecology of urban Norway and black rats (Rattus norvegicus and Rattus rattus). Urban Ecosyst. 2014;17:149–62.
Himsworth CG, Parsons KL, Jardine C, Patrick DM. Rats, cities, people, and pathogens: a systematic review and narrative synthesis of literature regarding the ecology of rat-associated zoonoses in urban centers. Vector Borne Zoonotic Dis. 2013;13:349–59.
Beaucournu JC, Moreno L, González-Acuña D. Le genre Dasypsyllus Baker 1905: description d’une espèce nouvelle et essai de mise au point sur ce genre (Siphonaptera: Ceratophyllidae). Ann la Soc Entomol Fr. 2016;51:470–6.
Beaucournu JC, Moreno L, González-Acuña D. Fleas (Insecta-Siphonaptera) of Chile: a review. Zootaxa. 2014;3900:151–203.
Labruna MB, Ogrzewalska M, Moraes-Filho J, Lepe P, Gallegos JL, López J. Rickettsia felis in Chile. Emerg Infect Dis. 2007;13:1794–5.
Poo-Muñoz DA, Elizondo Patrone MC, Escobar LE, Astorga F, Bermúdez S, Martinez-Valdebenito CP, et al. Fleas and ticks in carnivores from a domestic-wildlife interface: implications for public health and wildlife. J Med Entomol. 2016;53:1433–43.
Müller A, Rodríguez E, Walker R, Bittencourt P, Pérez-Macchi S, Ricardo Gonçalves L, et al. Occurrence and genetic diversity of Bartonella spp. (Rhizobiales: Bartonellaceae) and Rickettsia spp. (Rickettsiales: Rickettsiaceae) in cat fleas (Siphonaptera: Pulicidae) from Chile. J Med Entomol. 2013;142:1627–32.
Cevidanes A, Di Cataldo S, Vera F, Lillo P, Millán J. Molecular detection of vector-borne pathogens in rural dogs and associated Ctenocephalides felis fleas (Siphonaptera: Pulicidae) in Easter Island (Chile). J Med Entomol. 2018;55:1659–63.
Moreno L, Espinoza-Carniglia M, Lizama-Schmeisser N, Torres LG, Silva-De La Fuente MC, Lareschi M, et al. Fleas of black rats (Rattus rattus) as reservoir host of Bartonella spp. in Chile. PeerJ. 2019;7:e7371.
Meerburg BG, Singleton GR, Kijlstra A. Rodent-borne diseases and their risks for public health rodent-borne diseases and their risks for public health. Crit Rev Microbiol. 2009;35:221–70.
Instituto Nacional de Estadisticas (Chile). Ciudades, pueblos, aldeas y caseríos. 2005. http://arks.princeton.edu/ark:/88435/dsp019g54xm55n. Accessed 22 Apr 2015.
Mills JN, Yates TL, Childs JE, Parmenter RR, Ksiazek TG, Rollin PE, et al. Guidelines for working with rodents potentially infected with Hantavirus. J Mammal. 1995;76:716.
Carpenter J, Marion C. Exotic animal formulary. 5th ed. Missouri: Elsevier Saunders Press; 2018.
Iriarte A. Los mamíferos de Chile. Santiago: Lynx edition; 2008.
Labruna MB, Whitworth T, Horta MC, Bouyer DH, McBride JW, Pinter A, et al. Rickettsia species infecting Amblyomma cooperi ticks from an area in the state of São Paulo, Brazil, where brazilian spotted fever is endemic. J Clin Microbiol. 2004;42:90–8.
Roux V, Raoult D. Phylogenetic analysis of members of the genus Rickettsia using the gene encoding the outer-membrane protein rOmpB (ompB). Int J Syst Evol Microbiol. 2000;50:1449–55.
Johnson P. A classification of the Siphonaptera of South America, with descriptions of new species. Washington: Entomological Society of Washington; 1957.
Sánchez J, Lareschi M. Two new species of Neotyphloceras (Siphonaptera: Ctenophthalmidae) from Argentinean Patagonia. Zootaxa. 2014;3784:159–70.
Lewis RE. Résumé of the Siphonaptera (Insecta) of the World. J Med Entomol. 1998;35:377–89.
Duh D, Punda-Polic V, Avsic-Zupanc T, Bouyer D, Walker DH, Popov VL, et al. Rickettsia hoogstraalii sp. nov., isolated from hard-and soft-bodied ticks. Int J Syst Evol Microbiol. 2010;60:977–84.
Loyola S, Flores-Mendoza C, Torre A, Kocher C, Melendrez M, Luce-Fedrow A, et al. Rickettsia asembonensis characterization by multilocus sequence typing of complete genes, Peru. Emerg Infect Dis. 2018;24:931–3.
Ogata H, La Scola B, Audic S, Renesto P, Blanc G, Robert C, et al. Genome sequence of Rickettsia bellii illuminates the role of amoebae in gene exchanges between intracellular pathogens. PLoS Genet. 2006;2:733–44.
Medina-Pinto RA, Torres-Castro MA, Medina-Pinto RA, Bolio-González ME, Rodríguez-Vivas RI. Natural cysticercus fasciolaris infection in rodents from a rural area in Yucatan, Mexico. Vet Mex. 2019;6:1–10.
Whisson DA, Quinn JH, Collins KC. Home range and movements of roof rats (Rattus rattus) in an old-growth riparian forest, California. J Mammal. 2007;88:589–94.
Ahmed S, Dávila JD, Allen A, Tacoli And C, Fèvre EM. Does urbanization make emergence of zoonosis more likely? Evidence, myths and gaps. Environ Urban. 2019;31:443–60.
Monteverde Hadora. Movimientos de roedores intra- e inter-ambiente y riesgo de exposición al Hantavirus “Andes” en Patagonia norte, Argentina. Ecol Austral. 2017;27:279–89.
Caron A, Cappelle J, Cumming GS, de Garine-Wichatitsky M, Gaidet N. Bridge hosts, a missing link for disease ecology in multi-host systems. Vet Res. 2015;46:83.
Radzijevskaja J, Kaminskiene E, Lipatova I, Mardosaite-Busaitiene D, Balčiauskas L, Stanko M, et al. Prevalence and diversity of Rickettsia species in ectoparasites collected from small rodents in Lithuania. Parasit Vectors. 2018;11:375.
Kuo CC, Huang JL, Lin TE, Wang HC. Detection of Rickettsia spp and host and habitat associations of fleas (Siphonaptera) in eastern Taiwan. Med Vet Entomol. 2012;26:341–50.
Kim HC, Yang YC, Chong ST, Ko SJ, Lee SE, Klein TA, et al. Detection of Rickettsia typhi and seasonal prevalence of fleas collected from small mammals in the republic of Korea. J Wildl Dis. 2010;46:165–72.
Merhej V, Raoult D. Rickettsial evolution in the light of comparative genomics. Biol Rev. 2011;86:379–405.
Krawczak FS, Labruna MB, Hecht JA, Paddock CD, Karpathy SE. Genotypic characterization of Rickettsia bellii reveals distinct lineages in the United States and South America. Biomed Res Int. 2018;2018:8505483.
Parola P, Paddock CD, Socolovschi C, Labruna MB, Mediannikov O, Kernif T, et al. Update on tick-borne rickettsioses around the world: a geographic approach. Clin Microbiol Rev. 2013;26:657–702.
Fortes FS, Silveira I, Moraes-Filho J, Leite RV, Bonacim JE, Biondo AW, et al. Seroprevalence of Rickettsia bellii and Rickettsia felis in dogs, São José dos Pinhais, State of Paraná, Brazil. Rev Bras Parasitol Vet. 2010;19:222–7.
Horta MC, Sabatini GS, Moraes-Filho J, Ogrzewalska M, Canal RB, Pacheco RC, et al. Experimental infection of the opossum Didelphis aurita by Rickettsia felis, Rickettsia bellii, and Rickettsia parkeri and evaluation of the transmission of the infection to ticks Amblyomma cajennense and Amblyomma dubitatum. Vector Borne Zoonotic Dis. 2010;10:959–67.
Song S, Chen C, Yang M, Zhao S, Wang B, Hornok S, et al. Diversity of Rickettsia species in border regions of northwestern China. Parasit Vectors. 2018;11:634.
Robinson MT, Satjanadumrong J, Hughes T, Stenos J, Blacksell SD. Diagnosis of spotted fever group Rickettsia infections: the Asian perspective. Epidemiol Infect. 2019;147:e286.
Karasartova D, Gureser AS, Gokce T, Celebi B, Yapar D, Keskin A, et al. Bacterial and protozoal pathogens found in ticks collected from humans in Corum province of Turkey. PLoS Negl Trop Dis. 2018;12:e0006395.
Brinkmann A, Hekimoǧlu O, Dinçer E, Hagedorn P, Nitsche A, Ergünay K. A cross-sectional screening by next-generation sequencing reveals Rickettsia, Coxiella, Francisella, Borrelia, Babesia, Theileria and Hemolivia species in ticks from Anatolia. Parasit Vectors. 2019;12:26.
Chisu V, Leulmi H, Masala G, Piredda M, Foxi C, Parola P. Detection of Rickettsia hoogstraalii, Rickettsia helvetica, Rickettsia massiliae, Rickettsia slovaca and Rickettsia aeschlimannii in ticks from Sardinia. Italy. Ticks Tick Borne Dis. 2017;8:347–52.
Maina AN, Jiang J, Luce-Fedrow A, St. John HK, Farris CM, Richards AL. Worldwide presence and features of flea-borne Rickettsia asembonensis. Front Vet Sci. 2019;5:334.
Jiang J, Maina AN, Knobel DL, Cleaveland S, Laudisoit A, Wamburu K, et al. Molecular detection of Rickettsia felis and Candidatus Rickettsia asembonensis in fleas from human habitats, Asembo, Kenya. Vector Borne Zoonotic Dis. 2013;13:550–8.
Luce-Fedrow A, Maina AN, Otiang E, Ade F, Omulo S, Ogola E, et al. Isolation of Candidatus Rickettsia asembonensis from Ctenocephalides fleas. Vector Borne Zoonotic Dis. 2015;15:268–77.
Kocher C, Morrison AC, Leguia M, Loyola S, Castillo RM, Galvez HA, et al. Rickettsial disease in the Peruvian Amazon basin. PLoS Negl Trop Dis. 2016;10:e0004843.
Troyo A, Moreira-Soto RD, Calderon-Arguedas Ó, Mata-Somarribas C, Ortiz-Tello J, Barbieri ARM, et al. Detection of rickettsiae in fleas and ticks from areas of Costa Rica with history of spotted fever group rickettsioses. Ticks Tick Borne Dis. 2016;7:1128–34.
Dall’Agnol B, Souza U, Webster A, Weck B, Stenzel B, Labruna M, et al. “Candidatus Rickettsia asembonensis” in Rhipicephalus sanguineus ticks. Brazil. Acta Trop. 2017;167:18–20.
Tay ST, Koh FX, Kho KL, Sitam FT. Rickettsial infections in monkeys. Malaysia. Emerg Infect Dis. 2015;21:545–7.
Kolo AO, Sibeko-Matjila KP, Maina AN, Richards AL, Knobel DL, Matjila PT. Molecular detection of zoonotic Rickettsiae and Anaplasma spp. in domestic dogs and their ectoparasites in Bushbuckridge, South Africa. Vector Borne Zoonotic Dis. 2016;16:245–52.
Stevenson HL, Labruna MB, Montenieri JA, Kosoy MY, Gage KL, Walker DH. Detection of Rickettsia felis in a New World flea species, Anomiopsyllus nudata (Siphonaptera: Ctenophthalmidae). J Med Entomol. 2005;42:163–7.
Panti-May JA, Torres-Castro M, Hernández-Betancourt S, Dzul-Rosado K, Zavala-Castro J, López-Avila K, et al. Detection of Rickettsia felis in wild mammals from three municipalities in Yucatan, Mexico. Ecohealth. 2015;12:523–7.
Dieme C, Bechah Y, Socolovschi C, Audoly G, Berenger JM, Faye O, et al. Transmission potential of Rickettsia felis infection by Anopheles gambiae mosquitoes. Proc Natl Acad Sci USA. 2015;112:8088–93.
Acknowledgements
We thank the support provided by the Servicio Agrícola y Ganadero (SAG) and Corporación Nacional Forestal (CONAF). English-language editing of this manuscript was provided by Journal Prep Services, Toronto, Canada.
Funding
This study was supported by the National Fund for Scientific and Technological Development (FONDECYT 11150875, 1130948 and 1170972).
Author information
Authors and Affiliations
Contributions
LMS, MEC, NLS, LGTF, MCSF and DGA participated in rodent sampling, identifying rodent species and extracting the fleas. LMS, NLS and ML identified the fleas in the laboratory. MEC performed DNA extraction, PCR analyses and analyzed the data. LMS analyzed the data, wrote and edited the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All procedures were approved by the Universidad de Concepción bioethics committee, Servicio Agrícola y Ganadero (SAG R.E: 8968-2015, 1657.2016, 73-2016, 23-2017), and Corporación Nacional Forestal (CONAF N°018-2015).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Moreno-Salas, L., Espinoza-Carniglia, M., Lizama-Schmeisser, N. et al. Molecular detection of Rickettsia in fleas from micromammals in Chile. Parasites Vectors 13, 523 (2020). https://doi.org/10.1186/s13071-020-04388-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-020-04388-5