Abstract
Introduction
To date, a total of 2574 validated flea species have been discovered. Vermipsyllidae is a family of fleas that comprises at least eight species. Vermipsylla is a genus of the family Vermipsyllidae within the order Siphonaptera of fleas. Here a novel Vermipsylla species was described, and rickettsial agent was also detected in it.
Methods
A total of 128 fleas were collected directly from 260 pastured sheep in China. Of these, eight representative fleas (four males and four females) were identified by key morphological features. Meanwhile, 120 flea DNAs, including six flea samples for molecular taxonomy, were subjected to Rickettsia spp. DNA detection. The molecular identity of fleas was determined by amplification and sequenmce analysis of four genetic markers (the 28S rDNA genes, the 18S rDNA genes, the mitochondrial cytochrome c oxidase subunit I and subunit II). In addition, five Rickettsia-specific gene fragments were used to identify the species of the rickettsial agents. The amplified products were sequenced and phylogenetically analyzed.
Results
The morphological characteristics of the flea species identified in this study were similar to Vermipsylla alakurt, but presented difference in hair number of the metepimeron, the third tergum, the genitals and the tibiae of hind leg. The 18S rDNA, 28S rDNA and COII genetic markers from fleas showed the highest identity to those of V. alakurt, shared 98.45% (954/969), 95.81% (892/931) and 85.86% (571/665) similarities, respectively. However, the COI sequence showed the highest identity to that of Dorcadia ioffi with 88.48% (576/651) similarity. Rickettsia raoutii tested positive in 14.17% (17/120) flea DNA samples.
Conclusion
Our study reports the detection of R. raoultii in V. alakurt-like fleas infesting sheep in China.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Fleas (Insecta: Siphonaptera) are small, laterally flattened, wingless, and highly specialised insects [1], which belong to arthropod phylum, insecta, Siphonaptera. Currently, at least 2575 validated flea species belonging to 16 families and 246 genera have been described [2]. Vermipsylla, a genus of the family Vermipsyllidae, includes eight validated species, i.e. Vermipsylla alakurt (Kazakhstan, Mongolia, China), V. asymmetrica (China), V. ibexa (China), V. minuta (China), V. parallela (China), V. perplexa (China, Nepal), V. quilianensis (China) and V. yeae (China) [3, 4]. V. alakurt was firstly identified in China in 1965, in the southern region of Xinjiang Uygur Autonomous Region (XUAR), northwestern China [5]. During December to March, the adult fleas are mainly endemic in alpine pastoral areas, and prevailingly infest sheep, yaks, horses, yellow cattle and some wildlife species, causing irritation, poor condition, anaemia, abortion and even death [6,7,8].
Fleas are of tremendous medical and economic importance as vectors of several diseases important to human health including bubonic plague, murine typhus, and epidemic typhus [9, 10]. Rickettsia typhi in Ctenocephalides felis, Rickettsia felis in Liposcelis bostrychophila and Candidatus Rickettsia barbariae in V. alakurt, were previously reported [11,12,13].
Materials and Methods
Sample Collection and Identification of Fleas
In January 2018, fleas (128 in total) were collected directly from the entire body of 260 pastured sheep from two sheep flocks in Altaw Mountain, Wenquan County (the north region of XUAR, 2200 m a.s.l; 44°470ʹ30 N, 80°53ʹ30 E), which was adjacent to Kazakhstan [14]. The collected fleas were divided into two parts. Eight representative fleas (four males and four females) were for morphological identification by Stereomicroscope according to key features [13, 15] (eg. body length, labial palpus and notch of the tibiae of hind leg). In addition, DNAs of the individual fleas were extracted using the TIANamp Genomic DNA Kit (TIANGEN, Beijing, China) according to the manufacturer’s instructions. Six DNA from six representative flea samples were subjected to PCR amplification of four genetic markers [the 28S rDNA gene, the 18S rDNA gene and the mitochondrial cytochrome c oxidase subunit I (COI) and subunit II (COII)] for molecular identification. The nucleotide sequence were manually edited and compared to GenBank reference sequences (http://www.ncbi.nlm.nih.gov/BLAST/). Phylogenetic trees were constructed by the MEGA 7.0 software with the Maximum Likelihood (ML) method [16].
Detection of Rickettsial Agents and Sequence Analysis
A total of 120 flea DNAs, including six flea samples for molecular taxonomy, were subjected to PCR amplification for the detection of Rickettsia spp. DNA. Five rickettsial genetic markers, 17-kilodalton antigen 17-kilodalton antigen (17-kDa), surface cell antigen 4 (sca4), citrate synthetase (gltA), surface cell antigen 1 (sca1), and outer membrane proteins A (ompA) were used according to published protocols [17, 18]. Each PCR assay included a negative control (distilled water instead of flea DNA template) and a positive control (DNA from Candidatus R. barbariae obtained from V. alakurt). The above procedures were applied to treat the PCR products and their corresponding sequences. A phylogenetic tree was constructed by the MEGA 7.0 software with the ML method [13].
Results
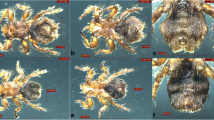
A total of 128 fleas were collected from the entire body of 260 sheep (10 males, 250 females) in two flocks in Altaw Mountain, Wenquan County, which is adjacent to Kazakhstan [14]. The fleas were divided into two parts: eight fleas (four males and four females) were preserved for morphological identification, and the remains were used for other purposes. The collected fleas had the following morphological characteristics similar to V. alakurt, which can be clearly distinguished from the other seven Vermipsylla species (shown in Table 1). Its size was the largest in the members of Vermipsylla genus, with males measuring 3.7–4.9 mm and females measuring 5.6–7.5 mm or longer. Its labial palpus was no longer than 17 segments, with 12–15 segments in females and 10–14 segments in males. The rear part of the female flea was full of fat, making up about three-quarters of its body length. The head of the intromittent organ of the males looked like a winter glove (with the back four fingers held together). The head of the spermathecae is ellipsoid, and the tail part was thin and long, with a sausage-like shape. The above morphological characteristics was similar to V. alakurt, interestingly, the females had less hairs (n = 5) next to the notch of the tibiae of hind leg, which obviously distinguishes from V. alakurt (n = 13). The differences from other key morphological features, eg. the hair of the metepimeron, the hair of the third tergum, the hair of the tibiae of hind leg and the hair of the genitals, were shown in Fig. 1 and Table 2. Due to the limited data available in GenBank, only two Vermipsyllidae species, V. alakurt and Dorcadia ioffi, were compared. The sequenced PCR product of 18S rDNA, 28S rDNA and COII obtained from fleas in this study were 98.45% (954/969), 95.81% (892/931) and 85.86% (571/665) similarities to V. alakurt (GenBank accession number KR297206, KR297207, KT193612), respectively. However, the COI nucleotide sequence of fleas in this study were 91.16% identical to D. ioffi in GenBank (accession number MF124314). Based on morphological and molecular evidence, all the fleas were identified as a novel Vermipsylla species, here named as Vermipsylla alakurt-like.
Photomicrographs of V. alakurt-like and V. alakurt. 1a: the whole body of V. alakurt-like, female. 1b: the whole body of V. alakurt, female. 2a: the metepimeron of V. alakurt-like, female. 2b: the metepimeron of V. alakurt, female. 3a: the third tergum of V. alakurt-like, female. 3b: the third tergum of V. alakurt, female. 4a: the tibiae leg of V. alakurt-like, female. 4b: the tibiae leg of V. alakurt, female. 5a: the spermathecae of V. alakurt-like, female. 5b: the spermathecae of V. alakurt, female. 6a: the whole body of V. alakurt-like, male. 6b: the whole body of V. alakurt, male. 7a: the metepimeron of V. alakurt-like, male.7b: the metepimeron of V. alakurt, male. 8a: the third and fourth tergum of V. alakurt-like, male. 8b: the third and fourth tergum of V. alakurt, male. 9a: the tibiae leg of V. alakurt-like, male. 9b: the tibiae leg of V. alakurt, male. 10a: the aedoeagus of V. alakurt-like, male. 10b: the aedoeagus of V. alakurt, male
Seventeen out of 120 flea DNA samples screened for Rickettsia spp. DNA were positive for the five genetic markers (17-kDa, ompA, sca4, gltA and sca1). No nucleic acids were amplified from the negative controls. The six responsive genetic markers, namely 17-kDa, gltA, ompA, sca4, and sca1 exhibited high sequence similarities with the genome of Rickettsia raoultii strain Khabarovsk (CP010969): 99.44% (360/362), 99.89% (896/897), 99.32% (441/444), 100% (762/762), and 99.64% (549/551), respectively. The phylogenetic tree revealed that R. raoultii was confirmed in this study (shown in Fig. 2).
Maximum-likelihood (ML; 500 bootstrap replicates) phylogenetic tree of the 17-kDa-ompA-sca4-gltA-sca1 constructed with MEGA7, using the sequences of R. raoultii from V. alakurt -like (◆) in this study and sequences from Rickettsia species retrieved from the GenBank database. The sequences for R. bellii were used as an outgroup. The scale bar represents the inferred substitutions per nucleotide site. The relative support for clades in the tree produced from the ML and NJ analyses are indicated above and below the branches, respectively
All of the obtained sequences were deposited in GenBank [17-kDa: MZ449221, gltA: OP376870, ompA: OP376871, sca4: OP376872, sca1: OP376873, 18S rDNA: OP339795, COI: OP324573, 28S rDNA: OP339754, COII: OP433454].
Discussion
Combined with molecular identity, systematic morphological identification in arthropods is important for the identification of a new species or subspecies [19, 20]. To date, morphology can also be used as a helpful tool to distinguish flea species or subspecies, such as C. felis and C. canis [21]. The flea species identified in this study can be classified into the Vermipsylla genus according to some morphological key features, such as no combs, its labial palpus less than 17 segments, the tibiae of hind leg having 6 notches, full of fat at the rear 3/4 part of the female body, only one seminal vesicle and its head being ellipsoid [22]. Although COI of Vermipsylla alakurt-like shared higher similarity (88.48%) with D. ioffi than datum between Vermipsylla alakurt-like and V. alakurt, we still believed Vermipsylla alakurt-like presents its own characteristics. Interestingly, Vermipsylla alakurt-like and V. alakurt have some similarities, such as segment number (n = 10–15) of labial palpus, the aedoeagus (in the shape of a winter glove) and the spermathecae (the head is ellipsoidal, and the tail is slender and sausage-shaped). Meanwhile, there are also some differences, especially in the number of hair and bristle of metepimeron, the third tergum, the tibiae of hind leg, aedoeagus and spermathecae (shown in Table 2), which makes it distinct from the other eight validated species of Vermipsylla genus, including V. alakurt. This finding indicates that the morphological characteristics (eg. the hair number of the metepimeron, the third tergum, the genitals and the tibiae of hind leg) could be helpful in identification and taxonomy especially for Vermipsylla species.
In this study, we firstly confirmed R. raoultii was detected in a novel Vermipsylla species. To date, R. raoultii have been detected in a sheep ked and 20 tick species, namely, Melophagus ovinus, Dermacentor nuttallii, De. marginatus, De. reticulatus, De. silvarum, Rhipicephalus pumilio, Rh. sanguineus, Rh. annulatus, Ixodes persulcatus, I. ricinus, I. canisuga, I. kaiseri, Haemaphysalis longicornis, Ha. erinacei, Ha. Punctata, Ha. concinna, Ha. japonica, Amblyomma helvolum, Hyalomma asiaticum, Hy. anatolicum and Hy. marginatum [23,24,25,26,27,28,29,30,31,32,33,34,35,36,37]. Previously, R. raoultii was rarely reported in fleas exception of C. felis [38]. This is the first time that R. raoultii has been found in Vermipsyllidae.
Although the vast of the spotted fever group rickettsiae (SFGR) are transmitted by ticks or sheep keds [23, 39], there are exceptions. R. africae, R. felis, Candidatus R. barbariae, belonging to the members of SFGR, were also detected in C. garei fleas (from passerine birds that had migrated from Africa) [40], C. felis fleas (collected from sheep, cats and dogs) [41] and V. alakurt (from sheep), respectively [13]. Herein, we reported the presence of R. raoultii in Vermipsylla alakurt-like fleas from sheep in an alpine pastoral area in the north-western of China. There is not enough evidence to confirm that Vermipsylla alakurt-like can transmit rickettsiosis to sheep. In future, detecting R. raoultii in sheep organs would demonstrate its potential to cause the disease.
Previously, the special geographical environment and current research confirms that R. raoultii is highly prevalent in the northwest of China and its neighboring countries, such as Mongolia, Russia and Kazakhstan [22, 42, 43]. We should pay attention to the transmission of SFGR in a wider arthropod species especially in the blood-suckers.
Conclusion
A novel Vermipsylla species from alpine pastoral region was described. This is the first report of the presence of R. raoultii in Vermipsyllidae. These findings extend our knowledge of Vermipsylla species, and the geographical distribution and reservoir hosts for R. raoultii. Future research should assess the vector competence and transmission dynamics of Vermipsylla alakurt-like for R. raoultii and other rickettsial pathogens.
Data Availability
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
References
Bitam I, Dittmar K, Parola P, Whiting MF, Raoult D (2010) Fleas and flea-borne diseases. Int J Infect Dis 14(8):e667–e676. https://doi.org/10.1016/j.ijid.2009.11.011
Zhu Q, Hastriter MW, Whiting MF, Dittmar K (2015) Fleas (Siphonaptera) are cretaceous, and evolved with theria. Mol Phylogenet Evol 90:129–139. https://doi.org/10.1016/j.ympev.2015.04.027
Lewis RE, Lewis JH (1994) Siphonaptera of North America north of Mexico: Vermipsyllidae and Rhopalopsyllidae. J Med Entomol 31(1):82–98. https://doi.org/10.1093/jmedent/31.1.82
Lewis RE (1973) Notes on the geographical distribution and host preferences in the order Siphonaptera: Part 2 Rhopalopsyllidae Malacopsyllidae and Vermipsyllidae. J Med Entomol 10(3):255–260. https://doi.org/10.1093/jmedent/10.3.255
Liu ZY, Wu HY, Wu FL (1965) On two new species of Vermipsylla from west China and a revision of the characters of the genus (Siphonaptera: Vermipsyllidae). Acta Zool Sinica 17(4):406–413
Wang Y, Wang GL, Li TW, Shayi PH, Li ZW, Li XY, Yasen KEB (2008) Research on Morphology of Vermipsylla alakurt of Animal. Grass-Feeding Livestock 03:40–41. https://doi.org/10.16863/j.cnki.1003-6377.2008.03.013
Wang GL, Xi N, Dang XS, Shi BX, Haxi BT, Sha WL (2004) Pathogen identification of Vermipsylla of domestic animal in Bazhou, Xinjiang. Chin J Anim Infect Dis 03:8–9+11
Wang GL, Xi N, Shawulie ADL, Simayi NYZ, Shi BX, Dang XS (2004) Studies on some characteristics of bioecology and morphology of Vermipsylla alakurt. Bull Dis Control Prevent China 03:25–27. https://doi.org/10.13215/j.cnki.jbyfkztb.2004.03.012
Galy A, Loubet P, Peiffer-Smadja N, Yazdanpanah Y (2018) The plague: an overview and hot topics. Rev Med Interne 39(11):863–868. https://doi.org/10.1016/j.revmed.2018.03.019
Woodward TE (1982) Murine and epidemic typhus rickettsiae: how close is their relationship? Yale J Biol Medb 55(3–4):335–341
Noden BH, Davidson S, Smith JL, Williams F (2017) First detection of Rickettsia typhi and Rickettsia felis in fleas collected from client-owned companion animals in the Southern Great Plains. J Med Entomol 54(4):1093–1097. https://doi.org/10.1093/jme/tjx069
Behar A, McCormick LJ, Perlman SJ (2010) Rickettsia felis infection in a common household insect pest, Liposcelis bostrychophila (Psocoptera: Liposcelidae). Appl Environ Microbiol 76(7):2280–2285. https://doi.org/10.1128/AEM.00026-10
Zhao SS, Li HY, Yin XP, Liu ZQ, Chen CF, Wang YZ (2016) First detection of Candidatus Rickettsia barbariae in the flea Vermipsylla alakurt from north-western China. Parasit Vectors 9(1):325–329. https://doi.org/10.1186/s13071-016-1614-2
Wang Y, Wang G, Cong PQ, Yu YW (2013) Study on lifecycle of Vermipsylla alakurt. Shandong J Anim Sci Vet Med 34:74–75
Wu HY et al (2007) Fauna Sinica Insecta Siphonaptera Second Edition (I). Science Press
Huang YL, Huang DN, Wu WH, Yang F, Zhang XM, Wang M, Tang YJ, Zhang Q, Peng LF, Zhang RL (2018) Identification and characterization of the causative triatomine bugs of anaphylactic shock in Zhanjiang. China Infect Dis Poverty 7(1):127–136. https://doi.org/10.1186/s40249-018-0509-1
Anstead CA, Chilton NB (2013) A novel Rickettsia species detected in Vole Ticks (Ixodes angustus) from Western Canada. Appl Environ Microbiol 79(24):7583–7589. https://doi.org/10.1128/AEM.02286-13
Spitalská E, Stefanidesová K, Kocianová E, Boldiš V (2012) Rickettsia slovaca and Rickettsia raoultii in Dermacentor marginatus and Dermacentor reticulatus ticks from Slovak Republic. Exp Appl Acarol 57(2):189–197. https://doi.org/10.1007/s10493-012-9539-8
Hornok S, Kontschán J, Kováts D, Kovács R, Angyal D, Görföl T, Polacsek Z, Kalmár Z, Mihalca AD (2014) Bat ticks revisited: Ixodes ariadnae sp. nov. and allopatric genotypes of I. vespertilionis in caves of Hungary. Parasit Vectors 7:202–210. https://doi.org/10.1186/1756-3305-7-202
Zurita A, Callejón R, García-Sánchez ÁM, Urdapilleta M, Lareschi M, Cutillas C (2019) Origin, evolution, phylogeny and taxonomy of Pulex irritans. Med Vet Entomol 33(2):296–311. https://doi.org/10.1111/mve.12365
García-Sánchez AM, Zurita A, Cutillas C (2022) Morphometrics as a complementary tool in the differentiation of two cosmopolitan flea species: Ctenocephalides felis and Ctenocephalides canis. Insects 13(8):707–715. https://doi.org/10.3390/insects13080707
Liu ZY (1986) Zoology of China: Insect class Siphonaptera. Science Press, Beijing, pp 258–294
Liu D, Wang YZ, Zhang H, Liu ZQ, Wureli HZ, Wang SW, Tu CC, Chen CF (2016) First report of Rickettsia raoultii and R. slovaca in Melophagus ovinus, the sheep ked. Parasit Vectors 9(1):600–605. https://doi.org/10.1186/s13071-016-1885-7
Olivieri E, Wijnveld M, Bonga M, Berger L, Manfredi MT, Veronesi F, Jongejan F (2018) Transmission of Rickettsia raoultii and Rickettsia massiliae DNA by Dermacentor reticulatus and Rhipicephalus sanguineus (s.l.) ticks during artificial feeding. Parasit Vectors 11(1):494–500. https://doi.org/10.1186/s13071-018-3075-2
Rydkina E, Roux V, Rudakov N, Gafarova M, Tarasevich I, Raoult D (1999) New Rickettsiae in ticks collected in territories of the former soviet union. Emerg Infect Dis 5(6):811–814. https://doi.org/10.3201/eid0506.990612
Hembram PK, Kumar GS, Kumar KGA, Deepa CK, Varghese A, Bora CAF, Nandini A, Malangmei L, Kurbet PS, Dinesh CN, Juliet S, Ghosh S, Ravindran R (2022) Molecular detection of pathogens in the ova and unfed larvae of Rhipicephalus annulatus and Haemaphysalis bispinosa ticks infesting domestic cattle of south India. Acta Trop 235:106656. https://doi.org/10.1016/j.actatropica.2022.106656
Liu G, Zhao SS, Tan WB, Hornok S, Yuan WM, Mi LG, Wang SW, Liu ZQ, Zhang YY, Hazihan WRLHZ, Gu XL, Wang YZ (2021) Rickettsiae in red fox (Vulpes vulpes), marbled polecat (Vormela peregusna) and their ticks in northwestern China. Parasit Vectors 14(1):204–209. https://doi.org/10.1186/s13071-021-04718-1
Zhao SS, Yang MH, Jiang MM, Yan B, Zhao SS, Yuan WM, Wang BJ, Hornok S, Wang YZ (2019) Rickettsia raoultii and Rickettsia sibirica in ticks from the long-tailed ground squirrel near the China-Kazakhstan border. Exp Appl Acarol 77(3):425–433. https://doi.org/10.1007/s10493-019-00349-5
Cheng CF, Fu WM, Ju WD, Yang LW, Xu N, Wang YM, Li H, Wang YL, Hu MX, Wen J, Jiao D, Geng C, Sun Y (2016) Diversity of spotted fever group Rickettsia infection in hard ticks from Suifenhe, Chinese-Russian border. Ticks Tick Borne Dis 7(5):715–719. https://doi.org/10.1016/j.ttbdis.2016.02.023
Stańczak J, Biernat B, Racewicz M, Zalewska M, Matyjasek A (2018) Prevalence of different Rickettsia spp. in Ixodes ricinus and Dermacentor reticulatus ticks (Acari: Ixodidae) in north-eastern Poland. Ticks Tick Borne Dis 9(2):427–434. https://doi.org/10.1016/j.ttbdis.2017.12.010
Seo MG, Kwon OD, Kwak D (2020) High prevalence of Rickettsia raoultii and associated pathogens in canine ticks. South Korea Emerg Infect Dis 26(10):2530–2532. https://doi.org/10.3201/eid2610.191649
Guo LP, Mu LM, Xu J, Jiang SH, Wang AD, Chen CF, Guo G, Zhang WJ, Wang YZ (2015) Rickettsia raoultii in Haemaphysalis erinacei from marbled polecats, China-Kazakhstan border. Parasit Vectors 8:461–463. https://doi.org/10.1186/s13071-015-1065-1
Doornbos K, Sumrandee C, Ruang-Areerate T, Baimai V, Trinachartvanit W, Ahantarig A (2013) Rickettsia sp. closely related to Rickettsia raoultii (Rickettsiales: Rickettsiaceae) in an Amblyomma helvolum (Acarina: Ixodidae) tick from a Varanus salvator (Squamata: Varanidae) in Thailand. J Med Entomol 50(1):217–220. https://doi.org/10.1603/me12010
Yin X, Guo S, Ding C, Cao M, Kawabata H, Sato K, Ando S, Fujita H, Kawamori F, Su H, Shimada M, Shimamura Y, Masuda S, Ohashi N (2018) Spotted fever group Rickettsiae in inner Mongolia, China, 2015–2016. Emerg Infect Dis 24(11):2105–2107. https://doi.org/10.3201/eid2411.162094
Li YC, Wen XX, Li M, Moumouni PFA, Galon EM, Guo QY, Rizk MA, Liu MM, Li J, Ji SW, Tumwebaze MA, Byamukama B, Chahan B, Xuan XN (2020) Molecular detection of tick-borne pathogens harbored by ticks collected from livestock in the Xinjiang Uygur Autonomous Region. China Ticks Tick Borne Dis 11(5):101478. https://doi.org/10.1016/j.ttbdis.2020.101478
Pereira A, Parreira R, Cotão AJ, Nunes M, Vieira ML, Azevedo F, Campino L, Maia C (2018) Tick-borne bacteria and protozoa detected in ticks collected from domestic animals and wildlife in central and southern Portugal. Ticks Tick Borne Dis 9(2):225–234. https://doi.org/10.1016/j.ttbdis.2017.09.008
Mediannikov O, Matsumoto K, Samoylenko I, Drancourt M, Roux V, Rydkina E, Davoust B, Tarasevich I, Brouqui P, Fournier PE (2008) Rickettsia raoultii sp. nov., a spotted fever group rickettsia associated with Dermacentor ticks in Europe and Russia. Int J Syst Evol Microbiol 58(Pt 7):1635–1639. https://doi.org/10.1099/ijs.0.64952-0
Güvendi M, Can H, Köseoğlu AE, Erkunt Alak S, Kandemir Ç, Taşkın T, Sürgeç E, Demir S, Değirmenci Döşkaya A, Karakavuk M, Gül A, Döşkaya M, Gürüz AY, Ün C (2022) Investigation of the genetic diversity and flea-borne pathogens in Ctenocephalides felis samples collected from goats in İzmir and Şanlıurfa provinces of Turkey. Comp Immunol Microbiol Infect Dis 90–91:101896. https://doi.org/10.1016/j.cimid.2022.101896
Sukhiashvili R, Zhgenti E, Khmaladze E, Burjanadze I, Imnadze P, Jiang J, St John H, Farris CM, Gallagher T, Obiso RJ, Richards AL (2020) Identification and distribution of nine tick-borne spotted fever group Rickettsiae in the Country of Georgia. Ticks Tick Borne Dis 11(5):101470. https://doi.org/10.1016/j.ttbdis.2020.101470
Sekeyová Z, Mediannikov O, Roux V, Subramanian G, Spitalská E, Kristofík J, Darolová A, Raoult D (2012) Identification of Rickettsia africae and Wolbachia sp. in Ceratophyllus garei fleas from Passerine birds migrated from Africa. Vector Borne Zoonotic Dis 12(7):539–543. https://doi.org/10.1089/vbz.2011.0645
Šlapeta J, Lawrence A, Reichel MP (2018) Cat fleas (Ctenocephalides felis) carrying Rickettsia felis and Bartonella species in Hong Kong. Parasitol Int 67(2):209–212. https://doi.org/10.1016/j.parint.2017.12.001
Speck S, Derschum H, Damdindorj T, Dashdavaa O, Jiang J, Kaysser P, Jigjav B, Nyamdorj E, Baatar U, Munkhbat E, Choijilsuren O, Gerelchuluun O, Römer A, Richards AL, Kiefer D, Scholz H, Wölfel R, Zöller L, Dobler G, Essbauer S (2012) Rickettsia raoultii, the predominant Rickettsia found in Mongolian Dermacentor nuttalli. Ticks Tick Borne Dis 3(4):227–231. https://doi.org/10.1016/j.ttbdis.2012.04.001
Turebekov N, Abdiyeva K, Yegemberdiyeva R, Dmitrovsky A, Yeraliyeva L, Shapiyeva Z, Amirbekov A, Oradova A, Kachiyeva Z, Ziyadina L, Hoelscher M, Froeschl G, Dobler G, Zinner J, Frey S, Essbauer S (2019) Prevalence of Rickettsia species in ticks including identification of unknown species in two regions in Kazakhstan. Parasit Vectors 12(1):197. https://doi.org/10.1186/s13071-019-3440-9
Funding
This research was supported in part by the National Key Research and Development Program (2022YFC2304004), National Natural Science Foundation of China (81960379), International Cooperation Projects of and Xinjiang Uygur Autonomous Region (2020E01035), Youth Innovation & Talent Cultivation Project of Shihezi University (CXPY202104) and High-Level Talent Initiative Foundation of Shihezi University (RCZK202033).
Author information
Authors and Affiliations
Contributions
LFS, TWB and WYZ conceived the research. LFS, ZSS, LET, XSS and WN conducted experiments. LFS, TWB and WYZ analysed the data and wrote the manuscript. All authors read and approved the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethics Approval and Consent to Participate
Not applicable.
Patient Consent for Publication
Written informed consent was obtained from the patient for publication of this case report and accompanying images.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, F., Zhao, S., Li, E. et al. Detection of Rickettsia raoultii in Vermipsylla alakurt-Like Fleas of Sheep in Northwestern China. Acta Parasit. 69, 776–784 (2024). https://doi.org/10.1007/s11686-024-00809-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11686-024-00809-y