Abstract
Background
Anaplasma marginale is an obligate intracellular bacterium and the main cause of bovine anaplasmosis in tropical and subtropical regions. In Egypt, data regarding the prevalence of A. marginale in ruminant hosts and of the circulating genotypes is lacking. This study therefore aimed to (i) investigate the presence, epidemiology and genotypes of A. marginale in cattle and buffaloes in Egypt, (ii) to evaluate suitable diagnostic tools and (iii) to identify co-infections of A. marginale with other selected tick-borne pathogens.
Methods
Blood samples were collected from 394 animals (309 cattle and 85 buffaloes) from three different areas in Egypt. For the detection of A. marginale infection, several tests were compared for their sensitivity and specificity: blood smear analysis, enzyme-linked immunosorbent assay (ELISA), PCR, real-time PCR and reverse line blot (RLB) assay. Co-infections with A. marginale, piroplasms and other Anaplasmataceae were surveyed by RLB while A. marginale genotypes were identified by amplifying and sequencing the partial msp1α gene.
Results
Anaplasma marginale DNA was amplified by qPCR in 68.3% of cattle and 29.4% of buffaloes. RLB showed infection with A. marginale in 50.2% of cattle and 42.5% of buffaloes. Blood smear analysis detected this agent in 16.2% of cattle and 2.4% of buffaloes. ELISA showed specific antibodies against A. marginale in 54.9% of cattle. Anaplasma marginale was associated, in cattle and buffaloes, with several tick-borne pathogens (Theileria annulata, Babesia bovis, Babesia bigemina, Babesia occultans and Anaplasma platys). A significant difference of A. marginale infection level was noticed in cattle, where animals between 3–5-years-old had a higher prevalence (79.2%) compared to those older than 5 years (36.4%) and younger than 3 years (59.7%) and one year (64.5%), respectively (P = 0.002281). Microsatellite analysis identified 15 different genotypes.
Conclusions
The epidemiological findings revealed high prevalence of A. marginale in cattle and buffaloes in all the investigated areas. The circulation of diverse genotypes was observed, most of these A. marginale genotypes being specific for Egypt. The qPCR assay was confirmed to be the most sensitive tool for detection of A. marginale in cattle and buffaloes even in the carrier state, highlighting the importance of using suitable diagnostic tests.

Similar content being viewed by others
Background
Tick-borne diseases (TBDs) are responsible for important health problems worldwide [1, 2]. In Egypt, TBDs cause major health disorders, in particular to exotic and cross-bred cattle, endangering the wellbeing of animals and the livelihood of their owners [3, 4]. Bovine theileriosis caused by Theileria annulata and bovine babesiosis caused by Babesia bovis and/or Babesia bigemina are the most common TBDs in Egypt [1, 5, 6]. They are among the main impediments of livestock production in Egypt due to the fact that both interfere with animal productivity [1, 5, 7, 8]. Bovine anaplasmosis is characterized by mild to severe hemolysis and anemia that adversely affects animal health, production and reproduction [9, 10]. It is caused by Anaplasma marginale, an intraerythrocytic rickettsia mainly transmitted by Rhipicephalus microplus ticks, but other tick species have also been incriminated as vectors worldwide [11, 12]. Mechanical transmission through contaminated needles or surgical instruments under poor hygienic conditions or through biting flies may also occur. Both tick and animal hosts are considered reservoirs for this pathogen and can become persistently infected with A. marginale. Co-infections with A. marginale and other tick-borne pathogens such as Theileria, Babesia and other Anaplasma species are common in cattle [4, 13,14,15].
The persistence of A. marginale infection is enabled by antigenic variation [9, 10, 16, 17]. The Major surface proteins (MSP) of A. marginale play an important role in the interaction with the host, as these are highly variable proteins and responsible for the invasion of host cells. This multigene protein family usually undergoes antigenic changes and the resulting amino acids (antigens) were found to be characteristic for each geographic area. In Egypt, data on the prevalence of A. marginale and the circulating genotypes are lacking. Furthermore, the tick species transmitting this pathogen are not fully characterized. However, we hypothesized that A. marginale is a major tick-borne pathogen in Egypt occurring frequently as a single infection or in co-infection with other pathogens in cattle and buffaloes.
This study therefore aimed to (i) investigate the presence, epidemiology and genotypes of A. marginale in cattle and buffaloes in Egypt, (ii) evaluate suitable diagnostic tools, and (iii) identify co-infections with A. marginale and other selected tick-borne pathogens.
Methods
Study areas
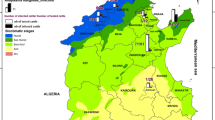
Samples were collected from cattle and buffaloes from three different regions in Egypt: Upper Egypt (EL-Minia and Assiut governorates), Middle Egypt (EL-Fayoum) and Egyptian oases (New Valley). Upper Egypt is a geographical and cultural division of Egypt running along the River Nile from Aswan northwards until the Delta. Middle Egypt is located between the Upper Egypt and Lower Egypt, from Assiut northwards until Memphis. The Egyptian oases is one of the biggest governorates in Egypt and represents more than 46% of the whole land area of Egypt. It is located at the plateau of the Egyptian western desert in southwestern Egypt and it borders Sudan to the south, River Nile to the east, and Libya to the west [18, 19] (Fig. 1).
Map of Egypt indicating the study areas. Cattle samples were collected from three different regions in Egypt: Upper Egypt (EL-Minia and Assiut governorates); Middle Egypt (El-Fayoum); and Egyptian oases (New Valley). Collection of samples from buffaloes was performed in the Upper Egypt (only in Assiut Governorate) and in Middle Egypt (EL-Fayoum)
Sample collection
Samples were collected from January to December 2018. Clinical examination was carried out on all animals before sampling. The examination included the measurement of body temperature, pulse and respiratory rate [20]. Three sample categories were collected from each animal: one blood sample in EDTA tubes from the ear vein for preparation of blood smears, one EDTA blood sample from the jugular vein for DNA extraction and another blood sample in a plain tube for serum preparation [21]. Inspection of the animal’s coats, udder, scrotum, inner side of the thighs and dewlap was performed for presence of ticks [20]. Tick samples were collected in 15 ml dry-screw cap Falcon™ tubes then transferred to 2 ml tubes containing 70% ethanol for preservation [22].
Tick identification
Ticks were identified to the species level using morphological identification keys under a ZEISS Stemi 508 stereomicroscope (Carl Zeiss, Oberkochen, Germany) [22, 23].
Microscopical examination
Thin blood smears were prepared and stained by Giemsa stain (Sigma-Aldrich, G4507, Darmstadt, Germany), then examined for the presence of blood pathogens under a light microscope (Olympus BX3M, Tokyo, Japan) using oil-immersion lens at a magnification of 1000× [21].
Serological diagnosis
Antibody detection was carried out by enzyme-linked immunosorbent assay (ELISA) on serum samples from cattle for detection of specific antibodies against A. marginale by using a commercial kit (SVANOVIR® A. marginale, Svanova, Uppsala, Sweden), according to the manufacturer’s instructions.
Molecular detection
DNA extraction
DNA extraction from blood of cattle and buffaloes was performed using the QIAamp DNA Blood kit (Qiagen, Hilden, Germany) according to the manufacturerʼs instructions.
Molecular detection of Anaplasma marginale by real time PCR
Anaplasma marginale was detected in cattle and buffalo samples by real time PCR (qPCR) targeting the msp1β gene [24]. The reaction was done in a total volume of 25 µl which included 12.5 µl (2×) iTaqTM Supermix (Bio-Rad Laboratories, Feldkirchen, Germany), 0.9 µl of molecular grade water, 0.5 µl of each primer (10 µM), 0.6 µl of the probe (5 µM) and 10 µl of DNA template. The qPCR program included an initial denaturation step at 95 ℃ for 10 min followed by 45 cycles of amplification at 95 ℃ for 45 s and 60 ℃ for 60 s then cooling. A positive control (A. marginale DNA from cattle, kindly provided by Dr Sándor Hornok), and a negative control (water) were added with each reaction.
Reverse line blot hybridization assay
Reverse line blot hybridization assay (RLB) was performed as previously described [13] for the simultaneous detection of several tick-borne pathogens including Theileria spp., Babesia spp., Anaplasma spp., Ehrlichia spp., Rickettsia spp. and Midichloria mitochondrii.
Conventional PCRs and sequencing
Anaplasmataceae
For confirmation of the RLB results, additional PCRs were performed to identify the Anaplasma species detected in some of the samples. A conventional PCR using GoTaq® Flexi DNA Polymerase kit (Promega, Madison, USA) was done to amplify a 855-bp fragment of the groEL gene of Anaplasma spp.
Since the GroEL PCR did not yield positive results in buffalo samples, a semi-nested PCR targeting the 16S rRNA gene of Anaplasma/Ehrlichia spp. was performed for the amplification of a fragment of 426 bp as described elsewhere [25] (Table 1). The PCR products were run on a 1.5% agarose gel stained with Roti®-Gel Stain Red (Carl Roth GmbH, Karlsruhe, Germany) for 40 min at 75 V and visualized with ChemiDocTM MP Imaging system (Bio-Rad Laboratories, Hercules, USA).
Babesia/Theileria species
Babesia and Theileria species were confirmed using two different primer pairs for each agent, targeting the 18S rRNA gene. Babesia spp. was detected using the previously described method by Casati et al. [26]. For detection of Theileria species, a Theileria-specific primer pair was used [27] (Table 1). The composition of the PCR mix and gel electrophoresis for both Babesia and Theileria were identical as described above for Anaplasma GroEL PCR.
Genotyping of Anaplasma marginale
A semi-nested PCR targeting the msp1α gene was carried out on 19 DNA samples from cattle and buffaloes that tested positive for A. marginale by qPCR, registering CT values ≤ 25 cycles [11, 16] (Table 1). The first PCR was done on both cattle and buffalo samples. Clear strong specific bands (size ranging from 800 to 1000 bp) were obtained from cattle samples while buffalo samples were subjected to the second amplification. The PCR reactions were performed using GoTaq® Flexi DNA Polymerase kit (Promega, Madison, USA) as previously described [11, 16, 17, 25, 28].
Sequencing
PCR sequencing reactions were performed for both forward and reverse strands. Reaction mix with a total volume of 10 µl included 1 µl 5× sequence buffer, 2 µl Big Dye ready for use master mix (Thermo Fischer, Darmstedt, Germany), 1 µl of each 10 µM primer and 5 µl molecular grade water. The thermal profile was 96 ℃ for 1 min as a primary denaturation, followed by 25 cycles of 96 ℃ for 10 s for denaturation, annealing temperature for 5 s depending on each used primer, 60 ℃ for 60 s for extension, and a final extension step at 72 ℃ for 5 min [29]. The PCR products were purified with NucleoSEQ® kit (Mackerey Nigel, Düren, Germany) according to the manufacturer’s instructions. After purification, 15 µl of each sample were mixed with 15 µl of the highly deionized (Hi-Di) formamide in 1.5 ml Eppendorf tube and sequenced on ABI PRISM® 3130 sequencer at the Institute of Diagnostic Virology, Friedrich-Loeffler-Institut, Germany.
Phylogenetic and microsatellite analysis of Anaplasma marginale msp1a gene
The obtained sequences were analyzed with Geneious 11.1.5 (https://www.geneious.com). The similarity search of the truncated sequences was carried out by using BLAST analysis (http://blast.ncbi.nlm.nih.gov/) after removal of the primer sequences. Nucleotide sequences of each sample were translated into amino acid sequences by the Open Reading Frame Finder translation tool on NCBI (https://www.ncbi.nlm.nih.gov/orffinder/). Nucleotide and protein sequences were aligned using the multiple-alignment program ClustalW [30]. The phylogenetic analysis was inferred using the Neighbor-Joining method [31]. The evolutionary distances were computed using the Kimura 2-parameter model [32]. Evolutionary analyses were conducted in MEGA X using A. phagocytophilum (HG528610) as the outgroup [33].
The obtained Egyptian A. marginale sequences were classified depending on the microsatellite (G/A TTT) m (GT) n in the 5’UTR region, located between the putative Shine-Dalgarno (SD) sequence (GTAGG) and the translation initiation codon (ATG). The SD-ATG distance was calculated in nucleotides as (4 × m) + (2 × n) + 1 [28, 34, 35].
Statistical analysis
Data were compared with Chi-square test using R in R Studio [36, 37]. Parameters related to animals such as age, breed and sex were investigated to find out the risk factors that may affect the animal susceptibility. In addition, some environmental factors like seasonal variation (hot months from April to September and non-hot months from October to March) and geographical areas (Upper Egypt, Middle Egypt and Egyptian oases) were taken in consideration during this study. The differences were considered significant at 5% threshold values. Detection tools for identification of A. marginale including blood smear, ELISA and RLB were evaluated against qPCR as a gold standard assay to calculate sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV) and combined predictive value (CPV) of each assay in both cattle and buffaloes [38].
Results
A total number of 309 cattle (140 males and 169 females) and 85 buffaloes (35 males and 50 females) were sampled during this study. Animals originated from three different localities: Middle Egypt (119 cattle and 23 buffaloes), Upper Egypt (111 cattle and 62 buffaloes) and Egyptian Oases (79 cattle). All examined animals were apparently healthy and were infested with adults (male and female) and nymphs of two tick species, Hyalomma excavatum and Rhipicephalus annulatus.
Anaplasma marginale was observed in 16.2% (50/309) and 2.4% (2/85) of the blood smears from cattle and buffalo, respectively (Fig. 2). Antibodies against A. marginale were detected in 54.8% (103/188) of the cattle serum samples. Infection rates of 68.3% (211/309) in cattle and 29.4% (25/85) in buffalo were found by qPCR. RLB showed infection with A. marginale in 50.2% (155/309) of cattle samples and 42.4% (36/85) of buffalo samples (Table 2). The RLB showed co-infections with A. marginale and other Anaplasma species and with piroplasms in both cattle and buffaloes. This was confirmed by sequencing of the RLB-PCR products. Co-infection with T. annulata was recorded in 49 (15.9%) cattle. Co-infection with B. bovis, B. bigemina and B. occultans was detected in 18 (5.8%), 2 (0.7%) and 1 (0.3%) cattle, respectively. Co-infection of A. marginale + A. platys was detected in 26 (8.4%) of the cattle samples. Buffaloes tested positive for co-infections with A. marginale and T. annulata (n = 1; 1.18%), B. bigemina (n = 2; 2.35%) and A. platys (n = 4; 4.71%).
The newly generated sequences from both cattle and buffalo were submitted to the GenBank database and are available under the following accession numbers: T. annulata: MN223723-MN223737; B. bigemina: MN227676-MN227679; B. occultans: MN227675; A. platys: MN202017-MN202023 and MN227688 and A. marginale: MN227687, MN227689-MN227692.
Real time PCR was considered as a gold standard detection assay in this study and according to the results obtained for A. marginale in both cattle and buffaloes, all detection assays were evaluated by assessing their sensitivity, specificity and predictive values (Table 3). The sensitivity of blood smear examination, ELISA and RLB assays in cattle was 23.7%, 66.7% and 60.7%, respectively, while in buffaloes the sensitivity of blood smears and RLB was 8.0% and 64.0%, respectively. On the other hand, the specificity of blood smear, ELISA and RLB in cattle was 100%, 78.0% and 84.7%, respectively, while in buffaloes the specificity of blood smear and RLB was 100% and 66.7%, respectively (Table 3).
Sex, breed or locality did not influence the prevalence of A. marginale. In addition, seasonal variation (hot months and non-hot months) also did not influence A. marginale prevalence, but it varied with age of the animal. There was a statistically significant difference according to age in cattle but not in buffaloes (Tables 4, 5).
Anaplasma marginale major surface protein α1 (msp1α) gene was sequenced for phylogenetic analysis and genotyping in both cattle and buffalo from different localities in Egypt. All sequences were also submitted to GenBank and can be retrieved using the following accession numbers: Middle Egypt (from cattle: MN273314, MN385279; from buffaloes: MN311245 and MN311246), Upper Egypt [Assiut governorate (from cattle: MN273312, MN273313 and MN385280; from buffaloes: MN 311243 and MN311244), EL-Minia governorate (MN257055: MN257057)] and Egyptian Oases (MN370071-MN370077).
The phylogenetic analysis revealed that the sequences for A. marginale group with different isolates from other countries or other local regions in Egypt (see Additional file 1: Figure S1).
Msp1α microsatellite sequences revealed that the isolates of A. marginale from Egypt are genetically different from other isolates reported worldwide and the microsatellite sequences produced SD-ATG distances between 23 and 27 nucleotides (Table 6). All the obtained sequences were translated into amino acids for assessing the tandems repeats (TR) in the Egyptian isolates. The results revealed that there are some amino acid repetitions specific for Egyptian A. marginale isolates. Twenty-seven TR sequences were found, 25 of which were identified for the first time in the Egyptian isolates and named as Eg1–Eg25. These TRs repeated between one to three times in each sample and resulted in the identification of 15 different genotypes in both cattle and buffaloes. The results also confirmed the occurrence of a predominant genotype in the collected samples with the following TRs sequence: Eg17, Eg23, Eg23, Eg23 and Eg24. This genotype was detected in cattle from two different areas including Upper Egypt (Assiut Governorate) and the Egyptian oases. On the other hand, two TRs (F) and (τ) matched with isolates from USA, Israel, Cuba and Brazil. The Egyptian isolate TR (F) is identical to the isolates from USA, Israel, Cuba and Brazil while the second isolate (τ) appeared with slight modification (Table 7).
Discussion
Anaplasma marginale is one of the most important tick-borne pathogens worldwide [39]. All animals investigated in this study were infested with ticks without showing clinical signs of anaplasmosis. The ticks were identified as Hyalomma excavatum and Rhipicephalus annulatus, both species being incriminated as vectors for A. marginale [40, 41].
The examination of Giemsa-stained blood smear was less sensitive compared to PCR-based detection methods potentially due to sampling subclinical or persistently infected animals that often show low numbers of infected erythrocytes. Another cause could be the dependency of the method for the microscopic visualization of A. marginale intraerythrocytic stage [42,43,44,45]. ELISA registered high sensitivity and low specificity compared to qPCR, being based on the detection of antibodies that occur during the persistent infection. Moreover, it depends on animal’s health and the ability of its immune system to produce antibodies against this variable antigenic pathogen.
Reverse line blot technique was highly sensitive and specific. Real time PCR was the most sensitive assay for detecting A. marginale. Previous studies registered similar findings, recommending molecular techniques for diagnosis of anaplasmosis [24]. These methods can overcome the persistent nature of the infection and the antigenic variability of the pathogens which adversely affect the ability of the serological tests to detect the infection [9, 24, 39].
Buffaloes in Middle and Upper Egypt showed lower infection rates compared to cattle from the same regions. These results could indicate a natural resistance against A. marginale in Egyptian buffaloes. Previous studies also stated that water buffaloes have the ability to reduce the infection and multiplication of this pathogen in the cells and also have natural resistance against tick infestation, reducing the probability of transmission of tick-borne pathogens [46,47,48]. Without clearing the persistent infection, buffaloes’ immune system can offer protection against the high level of rickettsemia and the acute phase of the disease if the animal is challenged with the homologous strain [49].
The prevalence of A. marginale was analyzed based on several risk factors related to host (age, sex, species and breed), environment, seasonal variation and study area. The sex of the animals did not affect the susceptibility. A study from Pakistan also did not find significant difference in the animals’ susceptibility according to their sex, but mainly linked with the degree of tick infestation. Animals exposed to heavy infestations were usually at higher risk than those exposed to light tick infestations [50]. Animal age was also among the factors affecting the probability of this infection in different animals. Older animals were more often infected than young ones. A previous study in Southern Queensland, also concluded that animals older than one year are more susceptible to the infection than younger animals [51]. On the contrary, a study in Pakistan concluded that animal susceptibility is not influenced by the age. Another study in Brazil concluded that animals below six months are more susceptible compared with older ones [12, 52]. Similar results to our findings regarding the cattle breeds were reported in Pakistan, confirming the lack of significance between infection rates of different cattle breeds [50]. Prevalence of A. marginale in Egypt was not affected by seasonal variation as suggested also in one study from Brazil [45] but in disagreement with a study in Southern Queensland which revealed that the disease is usually common during non-hot months (autumn and winter) [46]. The different localities did not affect the prevalence of A. marginale as in a previous study in Pakistan [50]. Based on these epidemiological findings and the genetic variability of A. marginale detected in different localities from previous studies [28, 39], we can conclude that prevalence and epidemiological features of A. marginale infection is closely related to its geographical distribution, each region having different genotypes.
Co-infections with A. marginale and pathogens such as T. annulata, B. bovis, B. bigemina, B. occultans and A. platys were found in both cattle and buffaloes. Anaplasma platys and B. occultans were detected for the first time in Egypt. Although A. platys is described as a tick-borne pathogen of dogs [53], it was previously also detected in cattle from China [54]. The co-infection with A. marginale and pathogens like Theileria spp., Babesia spp. and other Anaplasma species is a common finding and could be attributed to mixed infestation with several tick species or the ability of individual species to carry multiple pathogens [4, 13,14,15].
The phylogenetic analysis of A. marginale based on msp1α gene revealed that the Egyptian isolates were not only different from those circulating in other countries but also different from each other. The genetic differences found in the Egyptian isolates were independent of their locality. The occurrence of similar isolates in different localities may be a result of the uncontrolled movement of live animals between different localities in Egypt for marketing and slaughter. Although there is a marked difference between the Egyptian strains and other strains circulating worldwide, some strains are similar to isolates circulating in South Africa and Brazil perhaps due to importation of live animals from different countries to Egypt. Official data report for 2018 of the United States Department of Agriculture (USDA) stipulated that the importation of live animals is steadily increasing, and the same report revered that Sudan and Brazil are the largest suppliers for the live cattle.
The analysis of msp1α microsatellite sequences confirmed the presence of different genotypes amongst A. marginale strains obtained from both cattle and buffaloes from different localities in Egypt. Analysis of the translated sequences revealed that there were 25 new tandem repeats in the Egyptian isolates (Eg1 to Eg25), resulting in 15 different A. marginale genotypes. The two tandem repeats (F) and (τ) were previously isolated from USA, Israel, Cuba and Brazil, countries that act as suppliers of imported living animals to Egypt [55]. The Brazilian TR (τ) was found in cattle from Egyptian oases and the TR (F) from USA was found in cattle from Middle Egypt where there are some farms for the imported animals. These TRs found in different forms, include the original copy and another form with slight mutations that confirmed the occurrence of mutations to help the pathogens to adapt in the Egyptian field. In addition, the presence of predominant genotype, with the following TRs (Eg17, Eg23, Eg23, Eg23 and Eg24) in cattle from Upper Egypt (Assiut Governorate) and Egyptian oases (New Valley Governorate), is another evidence for the adverse effects of the uncontrolled animal movements which should be avoided by the veterinary authorities in Egypt. Water buffaloes are important hosts for A. marginale and can act as reservoir and transmit the infection to ticks. No genetic similarity was found in the msp1α gene structure of A. marginale isolated from cattle and water buffaloes except for one TR (Eg3) isolated from both species from Egyptian oases (MN370077, NV2). This finding could indicate that buffaloes act as reservoirs for specific genotypes which are not directly transmitted to cattle except after subjected to mutations. These mutations could make them infective and pathogenic for cattle especially in endemic areas and in mixed farms. The same hypothesis was indicated in previous studies stating that cattle may acquire A. marginale in superinfection with more than one genotype leading to generation of new genotypes. This process is sustained by the fact that antigenically, A. marginale is a highly variable pathogen, and these studies confirmed the use of this mechanism by A. marginale to escape the host immunity for persistence [56, 57].
Conclusions
In conclusion, infections with Anaplasma marginale were common in all the investigated areas from Egypt. Buffaloes were less often positive than cattle. Age of the animal was the main risk factor for positivity. Most isolates of A. marginale in Egypt were genetically diverse according to locality and host species. Anaplasma platys and B. occultans were detected in both cattle and buffaloes for the first time in Egypt. The qPCR and RLB assays are sensitive and specific for detection of A. marginale infection. Fifteen new Egyptian genotypes of A. marginale were identified in both cattle and buffaloes in Egypt.
Availability of data and materials
All data analyzed and generated during this study are included in this published article. The newly generated sequences were submitted to the GenBank database under the following accession numbers: 18S rRNA gene (T. annulata: MN223723-MN223737; B. bigemina: MN227676-MN227679; B. occultans: MN227675); GroEL/16S rRNA genes (A. platys: MN202017-MN202023 and MN227688); and MSP1 α gene (A. marginale: MN227687, MN227689-MN227692).
Abbreviations
- ELISA:
-
enzyme-linked immunosorbent assay
- PCR:
-
polymerase chain reaction
- qPCR:
-
real time PCR
- RLB:
-
reverse line blot
- msp1α:
-
major surface protein α1
- TRs:
-
tandem repetitions
- Hi-Di:
-
highly deionized formamide
- NCBI:
-
National Center for Biotechnology Information
- BLAST:
-
basic Local Alignment Search Tool
- SD:
-
putative Shine-Dalgarno
- Eg:
-
Egyptian genotype
- TP:
-
true positive
- TN:
-
true negative
- FP:
-
false positive
- FN:
-
false negative
- PPV:
-
positive predictive value
- NPV:
-
negative predictive value
- CPV:
-
combined predictive value
- USDA:
-
United States Department of Agriculture
References
AL-Hosary A, Ahmed L, Ahmed J, Nijhof A, Clausen P-H. Epidemiological study on tropical theileriosis (Theileria annulata infection) in the Egyptian Oases with special reference to the molecular characterization of Theileria spp. Ticks Tick-Borne Dis. 2018;9:1489–93.
Al-Hosary A, Ahmed L, Seitzer U. First report of molecular identification and characterization of Theileria spp. from water buffaloes (Bubalus bubalis) in Egypt. Adv Anim Vet Sci. 2015;3:629–33.
Abdel-Rady A, Ahmed LS, Mohamed A, Al-Hosary A. Using polymerase chain reaction (PCR) for diagnosis of bovine theileriosis in Upper Egypt. IJAVMS. 2010;4:67–74.
Awad H, Antunes S, Galindo RC, do Rosário VE, de la Fuente J, Domingos A, et al. Prevalence and genetic diversity of Babesia and Anaplasma species in cattle in Sudan. Vet Parasitol. 2011;181:146–52.
Al-Hosary AAT. Comparison between conventional and molecular methods for diagnosis of bovine babesiosis (Babesia bovis infection) in tick infested cattle in upper Egypt. J Parasit Dis Organ Indian Soc Parasitol. 2017;41:243–6.
Elhariri MD, Elhelw RA, Hamza DA, Soliman DE. Melecular detection of Anaplasma marginale in water buffaloes (Bubalus bubalis) based on majour surface protien. J Egypt Soc Parasitol. 2017;47:247–52.
Abdel-Rady A, Abd Ellah M, Kotb S. Clinical, diagnostic and therapeutic studies on theileriasis (Theileria annulata) in cattle in Upper Egypt. SCVMJ, XIII. 2008;2:387–95.
Elsify A, Sivakumar T, Nayel M, Salama A, Elkhtam A, Rizk M, et al. An epidemiological survey of bovine Babesia and Theileria parasites in cattle, buffaloes, and sheep in Egypt. Parasitol Int. 2015;64:79–85.
Kocan KM, de la Fuente J, Guglielmone AA, Meléndez RD. Antigens and alternatives for control of Anaplasma marginale infection in cattle. Clin Microbiol Rev. 2003;16:698–712.
Kocan KM, de la Fuente J, Blouin EF, Coetzee JF, Ewing SA. The natural history of Anaplasma marginale. Vet Parasitol. 2010;167:95–107.
Pothmann D, Poppert S, Rakotozandrindrainy R, Hogan B, Mastropaolo M, Thiel C, et al. Prevalence and genetic characterization of Anaplasma marginale in zebu cattle (Bos indicus) and their ticks (Amblyomma variegatum, Rhipicephalus microplus) from Madagascar. Ticks Tick Borne Dis. 2016;7:1116–23.
Rafique N, Kakar A, Iqbal A, Masood Z, Razzaq W, Iqbal F. Impact assessment of tick species, Rhipicephalus (Boophilus) microplus on the milk productions of cattle’s in the Quetta City of Province Balochistan, Pakistan. Global Veterinaria. 2015;15:19–23.
Hailemariam Z, Krücken J, Baumann M, Ahmed JS, Clausen P-H, Nijhof AM. Molecular detection of tick-borne pathogens in cattle from southwestern Ethiopia. PLoS ONE. 2017;12:e0188248.
Reinbold JB, Coetzee JF, Sirigireddy KR, Ganta RR. Detection of Anaplasma marginale and A phagocytophilum in bovine peripheral blood samples by duplex real-time reverse transcriptase PCR assay. J Clin Microbiol. 2012;50:284.
El-Ashker M, Hotzel H, Gwida M, El-Beskawy M, Silaghi C, Tomaso H. Molecular biological identification of Babesia, Theileria, and Anaplasma species in cattle in Egypt using PCR assays, gene sequence analysis and a novel DNA microarray. Vet Parasitol. 2015;207:329–34.
Lew AE, Bock RE, Minchin CM, Masaka S. A msp1 alpha polymerase chain reaction assay for specific detection and differentiation of Anaplasma marginale isolates. Vet Microbiol. 2002;86:325–35.
Almazán C, Alamzán C, Medrano C, Ortiz M, de la Fuente J. Genetic diversity of Anaplasma marginale strains from an outbreak of bovine anaplasmosis in an endemic area. Vet Parasitol. 2008;158:103–9.
Baines J, Malek J, Speake G. Cultural atlas of Ancient Egypt. New York: Checkmark Books; 2000.
Brugsch HK, Seymour HD, Smith P. A history of Egypt under the Pharaohs, derived entirely from the monuments: to which is added a memoir on the exodus of the Israelites and the Egyptian monuments; 2015. http:/core/books/history-of-egypt-under-the-pharaohs-derived-entirely-from-the-monuments/ECAA6635DB882E5F6850EB2810D2A381.
Radostitis O, Gay C, Blood DC, Hinchcliff KW. Veterinary medicine: a textbook of the diseases of cattle, horses, sheep, pigs and goats. 10th ed. Sydney: Saunders Ltd; 2006.
Coles EH. Veterinary clinical pathology. 4th ed. Philadelphia, London and Toronto: W. B. Saunders Company; 1986.
Estrada-Peña A, Bouattour A, Camicas JL, Walker AR. Ticks of domestic animals in the Mediterranean region. A guide of identification of species. Zaragoza, Spain: University of Zaragoza; 2004.
Hoogstraal H. Ticks of the Sudan (with special reference to Equatoria Province and with preliminary reviews of the genera Boophilus, Margaropus, and Hyallomma). Wash. 25: U.S Govt. Printing Office, D.C., USA. United States Naval Medical Research Unit No. 3; 1956.
Carelli G, Decaro N, Lorusso A, Elia G, Lorusso E, Mari V, et al. Detection and quantification of Anaplasma marginale DNA in blood samples of cattle by real-time PCR. Vet Microbiol. 2007;124:107–14.
Ybañez AP, Perez ZO, Gabotero SR, Yandug RT, Kotaro M, Inokuma H. First molecular detection of Ehrlichia canis and Anaplasma platys in ticks from dogs in Cebu, Philippines. Ticks Tick Borne Dis. 2012;3:288–93.
Casati S, Sager H, Gern L, Piffaretti J-C. Presence of potentially pathogenic Babesia sp. for human in Ixodes ricinus in Switzerland. Ann Agric Environ Med. 2017;13:65–70.
Sawczuk M, Maciejewska A, Skotarczak B. Identification and molecular characterization of Theileria sp. infecting red deer (Cervus elaphus) in northwestern Poland. Eur J Wildi Res. 2008;54:225–30.
de la Fuente J, Ruybal P, Mtshali MS, Naranjo V, Shuqing L, Mangold AJ, et al. Analysis of world strains of Anaplasma marginale using major surface protein 1a repeat sequences. Vet Microbiol. 2007;119:382–90.
Kieleczawa J, Mazaika E. Optimization of protocol for sequencing of difficult templates. J Biomol Tech. 2010;21:97–102.
Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80.
Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25.
Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–20.
Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–9.
Cabezas-Cruz A, Passos LMF, Lis K, Kenneil R, Valdés JJ, Ferrolho J, et al. Functional and immunological relevance of Anaplasma marginale major surface protein 1a sequence and structural analysis. PLoS One. 2013;8:e65243.
Estrada-Peña A, Naranjo V, Acevedo-Whitehouse K, Mangold AJ, Kocan KM, de la Fuente J. Phylogeographic analysis reveals association of tick-borne pathogen, Anaplasma marginale, MSP1a sequences with ecological traits affecting tick vector performance. BMC Biol. 2009;7:57.
R Core Development Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2019. https://www.R-project.org/.
RStudio Team. RStudio: integrated development for R. Boston: RStudio, Inc.; 2019. http://www.rstudio.com/.
Thrusfield M. Veterinary epidemiology. 3rd ed. New Jersey: Blackwell Science Publishing; 2007.
Palmer GH, Brown WC, Rurangirwa FR. Antigenic variation in the persistence and transmission of the ehrlichia Anaplasma marginale. Microbes Infect. 2000;2:167–76.
Samish M, Pipano E, Hadani A. Intrastadial and interstadial transmission of Anaplasma marginale by Boophilus annulatus ticks in cattle. Am J Vet Res. 1993;54:411–4.
Shkap V, Kocan K, Molad T, Mazuz M, Leibovich B, Krigel Y, et al. Experimental transmission of field Anaplasma marginale and the A. centrale vaccine strain by Hyalomma excavatum, Rhipicephalus sanguineus and Rhipicephalus (Boophilus) annulatus ticks. Vet Microbiol. 2009;134:254–60.
Eriks IS, Stiller D, Palmer GH. Impact of persistent Anaplasma marginale rickettsemia on tick infection and transmission. J Clin Microbiol. 1993;31:2091–6.
Kieser ST, Eriks IS, Palmer GH. Cyclic rickettsemia during persistent Anaplasma marginale infection of cattle. Infect Immun. 1990;58:1117–9.
Eriks IS, Palmer GH, McGuire TC, Allred DR, Barbet AF. Detection and quantitation of Anaplasma marginale in carrier cattle by using a nucleic acid probe. J Clin Microbiol. 1989;27:279–84.
Stich RW, Kocan KM, Palmer GH, Ewing SA, Hair JA, Barron SJ. Transstadial and attempted transovarial transmission of Anaplasma marginale by Dermacentor variabilis. Am J Vet Res. 1989;50:1377–80.
Aubry P, Geale DW. A review of bovine anaplasmosis. Transbound Emerg Dis. 2011;58:1–30.
Woolhouse MEJ, Webster JP, Domingo E, Charlesworth B, Levin BR. Biological and biomedical implications of the co-evolution of pathogens and their hosts. Nat Genet. 2002;32:569–77.
Obregón D, Corona BG, de la Fuente J, Cabezas-Cruz A, Gonçalves LR, Matos CA, et al. Molecular evidence of the reservoir competence of water buffalo (Bubalus bubalis) for Anaplasma marginale in Cuba. Vet Parasitol Reg Stud Rep. 2018;13:180–7.
Palmer GH, Rurangirwa FR, Kocan KM, Brown WC. Molecular basis for vaccine development against the ehrlichial pathogen Anaplasma marginale. Parasitol Today. 1999;15:281–6.
Rehman A, Conraths FJ, Sauter-Louis C, Krücken J, Nijhof AM. Epidemiology of tick-borne pathogens in the semi-arid and the arid agro-ecological zones of Punjab Province, Pakistan. Transbound Emerg Dis. 2019;66:526–36.
Rogers RJ, Blight GW, Knott SG. A study of the epidemiology of Anaplasma marginale infections of cattle in southern Queensland: clinical disease and the prevalence of complement fixing sntibodies. Aust Vet J. 1978;54:115–20.
Vieira LL, Canever MF, Cardozo LL, Cardoso CP, Herkenhoff ME, Neto AT, et al. Prevalence of Anaplasma marginale, Babesia bovis, and Babesia bigemina in cattle in the Campos de Lages region, Santa Catarina state, Brazil, estimated by multiplex-PCR. Parasite Epidemiol Control. 2019;6:e00114.
Harvey JW, Simpson CF, Gaskin JM. Cyclic thrombocytopenia induced by a Rickettsia-like agent in dogs. J Infect Dis. 1978;137:182–8.
Zhou Z, Li K, Sun Y, Shi J, Li H, Chen Y, et al. Molecular epidemiology and risk factors of Anaplasma spp., Babesia spp. and Theileria spp. infection in cattle in Chongqing, China. PLoS ONE. 2019;14:e0215585.
Abdi A, Roushdy S. Egyptian beef prices stable, consumption and imports to rise in 2019. USDA Foreign Agriculture Report, Livestock and Products Annual, approved by Minister-Counselor for Agricultural Affairs, Cairo, Egypt; 2018. p. 1–10.
Castañeda-Ortiz EJ, Ueti MW, Camacho-Nuez M, Mosqueda JJ, Mousel MR. Association of Anaplasma marginale strain superinfection with infection prevalence within tropical regions. PLoS ONE. 2015;10:e0129415.
Palmer GH, Brayton KA. Antigenic variation and transmission fitness as drivers of bacterial strain structure. Cell Microbiol. 2013;15:1969–75.
Acknowledgement
We would like to thank Dr Sándor Hornok, Department of Parasitology and Zoology, University of Veterinary Medicine, Budapest, Hungary, who supplied us with A. marginale DNA as a positive control. Further, we express our gratitude to Dr. Ahmed Makram Radwan (Jockey and Petzania Veterinary Clinics, Egypt) for his great help during this work.
Funding
Open Access funding enabled and organized by Projekt DEAL. The financial support was granted to AAH by Deutscher Akademischer Austauschdienst (DAAD) through GERSS program “German Egyptian Research Short term Scholarship GERSS” (57397533, 2018, Personal ID 91716072). Funding was also provided through DFG project “Molecular epidemiology network for promotion and support of delivery of life vaccines against Theileria parva and Theileria annulata infection in Eastern and Northern Africa” (DFG SE862/2-1).
Author information
Authors and Affiliations
Contributions
AAH and CS planned and coordinated the study. AAH performed field work. AAH, CR and OT performed the laboratory work. AAH and AMN performed the RLB. SF carried out the statistical analysis. All authors analyzed the data. AAH drafted the manuscript. CR, SF and CS critically revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Figure S1.
Phylogenetic tree inferred by using the Neighbor-Joining method, evolutionary distances were computed using the Kimura 2-parameter model in MEGA X and A. phagocytophilum (HG528610) as the outgroup.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
AL-Hosary, A., Răileanu, C., Tauchmann, O. et al. Epidemiology and genotyping of Anaplasma marginale and co-infection with piroplasms and other Anaplasmataceae in cattle and buffaloes from Egypt. Parasites Vectors 13, 495 (2020). https://doi.org/10.1186/s13071-020-04372-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-020-04372-z