Abstract
Background
Tick-borne diseases caused by Anaplasma species put serious constraints on the health and production of domestic cattle in tropical and sub-tropical regions. After recovering from a primary infection, cattle typically become persistent carriers of pathogens and play a critical role in the epidemiology of the disease, acting as reservoirs of the Anaplasma spp.
Methods
In this study a duplex PCR assay was used for the simultaneous detection of Anaplasma marginale and Anaplasma phagocytophilum in cattle using two primer pairs targeting msp4 and msp2 genes, respectively. We used this method to analyze DNA preparations derived from 328 blood cattle samples that were collected from 80 farms distributed among Tunisia’s four bioclimatic zones.
Results
The prevalence of the A. marginale infection (24.7 %) was significantly higher and more widespread (in all bioclimatic areas) than that of A. phagocytophilum (0.6 %), which was found in a mixed infection with A. marginale.
Conclusions
The duplex PCR assay used proved to be a rapid, specific and inexpensive mean for the simultaneous detection of Anaplasma marginale and Anaplasma phagocytophilum in cattle blood. It allowed us to report the identification of A. phagocytophilum for the first time in cattle in Tunisia and confirm the presence of A. marginale in cattle from several geographical areas of the country. Further epidemiological studies undertaken using this assay will help improve the surveillance of the associated diseases in the regions where they are endemic.
Similar content being viewed by others
Background
Among tick-borne diseases, bovine anaplasmosis is considered to be one of the most important in ruminants worldwide, causing significant economic losses in tropical and subtropical areas [1]. The socioeconomic impact of the disease and the restrictions on trading infected animals internationally led the Office International des Epizooties (OIE) Animal Health Code to categorize anaplasmosis as a disease that required a notification of its presence [2]. Because outbreaks are seasonal and infection rates are stable, the significance of anaplasmosis is underestimated in endemic areas [3]. Cattle can be infected by several Anaplasma species, like A. marginale, A. phagocytophilum, A. centrale and A. bovis [4–6]. Anaplasma marginale is one of the most prevalent tick-transmitted rickettsial diseases of cattle in the world [7]. Highly pathogenic, especially in cattle up to two years old, it causes a disease that produces progressive anemia and icterus [8]. Several decades ago A. phagocytophilum (formerly known as Ehrlichia phagocytophila, E. equi and human granulocytic ehrlichiosis agent), was identified in cattle; it may also infect humans [9]. Known to cause tick-borne fever in cattle, it causes not only high fever, but also coughs, miscarriages, decreased milk production and loss of appetite [10]. In areas infested by several tick vector species and where animal husbandry practices include vaccination with live A. centrale bacteria (Israel, Africa, Australia and parts of South America), cattle can be co-infected with two or more Anaplasma species [11, 12]. Disease treatment and prevention strategies focus on using reliable diagnostic tests to accurately and precisely identify infected cattle. While inoculating splenectomized cattle with whole blood has been the gold standard for determining persistent A. marginale infections in cattle, it is not required for routine testing [13]. Bovine anaplasmosis is diagnosed by identifying Anaplasma in Giemsa-stained blood smears from clinically suspect animals during the acute phase of the disease. However, this method is not useful for detecting pre-symptomatic and carrier animals. Currently, the competitive enzyme-linked immunosorbent assay (cELISA) is one of the most common diagnostic techniques used to identify the bovine anti-major surface protein 5 (anti-MSP5) of Anaplasma marginale [14]. It is considered to be a reliable screening test for cattle infected with A. marginale and to establish their carrier state. However, cross-reactivity has been reported when the cELISA is used to classify cattle infected with A. marginale and/or A. phagocytophilum [15, 16]. Several other serological tests have been used extensively in epidemiological studies of anaplasmosis despite the fact that they do not discriminate between different, antigenically similar Anaplasma species [16, 17]. Yet highly sensitive and specific, molecular methods have been developed to identify A. marginale and A. phagocytophilum DNA [18–22]. To develop a robust diagnostic method, an appropriate target needs to be selected in order to accurately and precisely determine an infection.
In Tunisia, Rickettsiales species including A. phagocytophilum, A. bovis, A. marginale, A. centrale, Ehrlichia canis, Ehrlichia sp. and A. platys have recently been detected in horses, cattle, small ruminants, camels, dogs and ticks [23–29]. A molecular assay based on a single-step duplex PCR, was used to simultaneously detect and differentiate A. marginale and A. phagocytophilum and determine their distribution in cattle from Tunisia.
Methods
Design of primers
A. marginale msp4 gene sequences and A. phagocytophilum msp2 gene sequences were aligned with those of other related species of the genera Anaplasma and Ehrlichia using Vector NTI 8.0 software (Informax Inc., North Bethesda, MD, US). Primers (Table 1) were designed to specifically amplify a 420 bp fragment of the msp4 gene of A. marginale and used in combination with the previously designed primer pair to amplify a 334 bp fragment of the msp2 gene of A. phagocytophilum [30].
Cloning and sequencing the msp4 A. marginale gene and msp2 A. phagocytophilum gene
DNA was extracted from whole blood samples of two cows naturally infected with A. marginale and A. phagocytophilum using QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany) as per the manufacturer’s recommendations, and extracted DNA was used as template to amplify a 420 bp (msp4 gene) and 334 bp (msp2 gene), respectively. The amplified products were cloned into a pCR4-TOPO vector and introduced into chemically competent Escherichia coli as per the manufacturer's instructions (TOPO TA cloning kit for sequencing; Invitrogen, Carlsbad, California). Recombinant plasmid DNA was purified using a FlexiPrep kit (Amersham Biosciences, Freiburg, Germany) and subjected to automatic dye terminator cycle sequencing. The nucleotide sequences of the plasmid inserts were confirmed as A. marginale and A. phagocytophilum by comparing them with the GenBank database.
The concentration of each plasmid was calculated with a NanoDrop® ND-1000 (Thermo Scientific, Wilmington, DE, USA) spectrophotometer and the plasmids were 10-fold serially diluted in a Tris-EDTA buffer to reach concentrations ranging from 108 to 10 copies/μl. Serial dilutions of individual plasmids as well as different combinations were tested to calculate the sensitivity of the assay.
Duplex amplification
PCR reactions were performed using a commercially available Multiplex-PCR assay kit (QIAGEN, Hilden, Germany) in 25 μl volume reactions that include 1× QIAGEN Multiplex PCR Master Mix (QIAGEN), 0.5 μM of Msp2-3 F/Msp2-3R primers, 0.2 μM M4-OvMar-F/M4-Mar-R primers and 5 μl of extracted DNA. Cycling conditions were 15 min at 95 °C, followed by 40 cycles of 94 °C for 30 s, 63 °C for 90 s, 72 °C for 90 s and 72 °C for 10 min. To avoid cross-contamination and false-positive reactions, we used plugged tips, set PCRs in separate rooms, and also included a negative (water) control in each run.
Sensitivity and specificity of single and duplex PCR assay
To determine the detection limit of single and duplex PCRs, 10-fold serial dilutions of individual plasmids with the insert of A. phagocytophilum and A. marginale as well as different combinations were tested under the conditions described above. Sensitivity was also tested on DNA extracted from blood from a non-infected cow spiked with these same plasmid combinations (Table 2). Specificity was tested using DNA from other species (Anaplasma ovis, Anaplasma platys, Ehrlichia sp., Ehrichia canis and Rickettsia conorii).
Study design and sampling approach
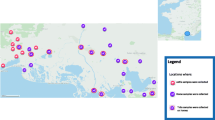
A cross-sectional study was carried out in 9 localities, located in 4 different bioclimatic zones, in northern and central Tunisia (humid, sub-humid, semi-arid and arid) where cattle’s breeding is an important economic activity (Fig. 1). All localities have a Mediterranean climate - cool, moist winters and dry, hot summers. Topographically, the areas have rolling hills interspersed with farmland, grassland, oak woodlands and Mediterranean scrub (Olea europaea, Pistacia lentiscus, Cistus monspeliensis, etc.). A total of 80 farms with fewer than 30 animals per farm were chosen randomly as representative of the local management system on the basis of the recommendations of the State Veterinary Office. Animal husbandry practices are generally traditional small herds grazing on permanent pastures or bush. A total of 328 cattle were sampled of which 37.2 % were local breed, 32.3 % cross-breeds, 18 % Friesian, 9.2 % Schwytz and 3.4 % Holstein. Animals ranged in age between 3 months and 13 years, and most were dairy cattle (97.6 %).
Blood sample collection and DNA extraction
Animals were bled once between June and November, a period during which they are typically grazing in pastures and exposed to tick bites. Blood was sampled in tubes containing ethylenediamine tetraacetic acid and DNA was extracted using the QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany). DNA yields were determined with a Nano-Drop® ND-1000 Spectrophotometer (Nano-Drop Technologies, DE, USA).
DNA samples were subjected to duplex PCR assay in order to detect A. marginale and A. phagocytophilum as described above, and amplicons were resolved in ethidium bromide-stained agarose (Gellyphor, EuroClone, Milan, Italy) gels (1.5 %) and measured by comparing them with the with Gene RulerTM 100-bp DNA Ladder (MBI Fermentas, Vilnius, Lithuania) as molecular marker. Gels were photographed using Gel Doc 2000 (Bio-Rad, Hercules, CA, USA).
Sequencing and data analysis
The specificity of the duplex PCR was confirmed by sequencing PCR amplicons of A. marginale and A. phagocytophilum using primers M4-OvMar-F/M4-Mar-R for msp4 gene and Msp2-3 F/Msp2-3R for msp2, respectively. Thirteen randomly chosen positive PCR products were purified using the ExoSAP cleanup procedure (Amersham Biosciences, Piscataway, NJ, USA). All nucleotide sequences were obtained using the Big Dye Terminator v.3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA) and the 3130 automated sequencer (Applied Biosystems). The sequences were edited and aligned using DNA Baser Sequence Aligner v3.5.4 software (Heracle BioSoft SRL, www.DnaBaser.com) to obtain optimal sequence alignment files. A BLAST analysis was made in the NCBI database to retrieve sets of homologues exhibiting high scores with the partial msp2 and msp4 gene of A. phagocytophilum and A. marginale, respectively.
Statistical analysis
The Chi-square or Fisher's exact tests were used to compare proportions of positivity in relation with bioclimatic zone, breed and sex. Observed differences were considered significant when the resulting P-value was less than 0.05.
Nucleotide sequence accession numbers
Sequence data were deposited in GenBank; accession numbers for the partial msp2 and msp4 sequences are KR871275–KR871287.
Results
Performance of the duplex PCR assay
Fragments of the expected size were generated from the template plasmids representing A. marginale (420 bp) and A. phagocytophilum (334 bp); while DNA from uninfected bovines used as negative control, displayed no evidence of fragment amplification. Similarly, no amplicons were obtained when testing DNA from other species (Anaplasma ovis, A. platys, Ehrlichia sp., E. canis and Rickettsia conorii). The PCR was able to detect 1 copy of A. marginale and 10 copies of A. phagocytophilum plasmid templates, when present as single infections, and 10 copies in mixed infections even when the differences in their concentrations were of two orders of magnitude and in the presence of uninfected host DNA (spiked controls).
Analysis of blood samples by PCR duplex assay
A total of 83 cattle (representing 25.3 % of the analyzed animals) were infected with A. marginale and/or A. phagocytophilum (Table 3). In 24.7 % (81/328) of analyzed cattle A. marginale msp4 amplicons were detected and, in another 0.6 % (2/328) of the animals, PCR results confirmed the presence of A. phagocytophilum DNA as a mixed infection with A. marginale (Table 3). None were positive for only A. phagocytophilum. The two cases of mixed infections were identified in the humid and sub-humid zones (Table 3), while A. marginale was detected with different rates in humid (25.6 %), sub-humid (46.6 %), semi-arid (8.8 %) and arid zones (4 %); the difference being significant (χ 2 = 47.95, df = 3, P < 0.0001) (Table 3). Infection rates of A. marginale were statistically higher in Schwyz breed (56.7 %) than in other breeds (χ 2 = 32.2, df = 4, P < 0.0001). The lowest prevalence was observed in Black Friesians (15.2 %; Table 4). Cattle with a mixed infection were local and Friesian black breeds (Table 4). Proportion of animals infected with A. marginale was significantly different (χ 2 = 7.22, df = 1, P = 0.0072) in cattle younger (2/34; 5.9 %) and older than (79/294; 26.9 %) one year. The two co-infected cattle were older than one year of age.
Sequence analyses
To confirm the PCR results, 11 PCR products positive for A. marginale (from 9 investigated localities) and two for A. phagocytophilum were sequenced. A BLAST analysis of the obtained sequences revealed genetic variability among A. marginale at five nucleotide positions (354, 423, 538, 564, 714) (Table 5). The 11 sequences (GenBank accession numbers KR871277–KR871287) showed significant identity (99–100 %) with A. marginale sequences described in Italy (GenBank accession number DQ000618), USA (GenBank accession number AY253143) and Spain (GenBank accession number AY456003). Four sequences (KR871279, KR871284, KR871281, KR871283) presented overlapping peaks at four positions. As they were confirmed by re-sequencing, they are indicative for double infections rather than for errors introduced during sequencing (Table 5). In addition, the two partial sequences of the msp2 gene of A. phagocytophilum (KR871275, KR871276) were identical between them and showed 100 % identity to sequences detected in white-tailed deer (DQ097228) and humans (AF135255) in America.
Discussion
Rickettsial (Anaplasmataceae) bacteria are recognized as emerging tick-borne pathogens that are important for humans and animals [31, 32]. Changes in the host-vector ecology are largely responsible for the emergence of these pathogens. Moreover, the new insights in the development of laboratory tools for the detection of Anaplasma infections have contributed to the detection of new species [33]. Indeed, the global threat of Anaplasmataceae highlights the need for new tools able to discriminate among the different species. Several molecular techniques were therefore proposed for detecting and characterizing species belonging to the family Anaplasmataceae. Most of these techniques target the major surface proteins (MSPs) [18], the heat-shock gene groEL [19], the 23S rRNA [20] and the 16S rRNA gene [21]. Here, we targeted the msp4 and msp2 genes, which are involved in host-pathogen and tick-pathogen interactions and have been used as markers for the genetic characterization of A. marginale strains [18], using a conventional PCR for the detection of A. phagocytophilum and A. marginale in a duplex format. The optimized duplex PCR was able to specifically detect A. phagocytophilum and A. marginale from both single and mixed DNA preparations without affecting the detection limit. The field samples provided further evidence of the assay’s applicability. Indeed, only amplicons of the expected size were generated and the results of the duplex PCR entirely corresponded with the results obtained by sequencing the amplicons generated. This duplex PCR allowed us to report the identification of A. phagocytophilum for the first time in cattle in Tunisia and confirm the presence of A. marginale in cattle from several geographical areas of the country.
The obtained prevalence of A. marginale (24.7 %) was lower than that reported in Kansas (37.6 %) [21], India (73.1–36.8 %) [34, 35], Sicily (50 %) [36], Brazil (70.2 %) [37], South African provinces (65–90 %) [38], Texas (82 %) [39] and Costa Rica (56.9 %) [40]. By contrast, this prevalence was higher than those recorded in Turkey (2.8 %) [41], Egypt (21.3 %) [42] and the Philippines (19.8 %) [43]. The significant prevalence of A. marginale warrants further investigation to evaluate the impact of this bacterium on livestock production, since it is considered to be a pathogenic species in cattle in North Africa, causing severe clinical symptoms and very serious economic losses [44]. However, at the time of blood sampling (June-November), the 83 cattle infected with A. marginale showed no clinical signs. These animals could be considered asymptomatic carriers. Indeed, Sergent et al. [44] have shown that North African strains of A. marginale confer an immune protection in experimentally infected animals.
Anaplasma marginale was identified in all the bioclimatic zones where we carried out our investigations, however, its prevalence was highest in farms in the sub-humid zone (46.6 %) compared to those in the humid (25.6 %), semi-arid (8.8 %) and arid (3.8 %) zones. These results concur with previous findings in Morocco [45], demonstrating a higher prevalence of infection with A. marginale in cattle in sub-humid (52 %) zones compared to humid (22.7 %) and semi-arid zone (20 %). The observed variations in the distribution of A. marginale in the different bioclimatic zones and farms in the same locality could be explained by the diversity of the Ixodidae fauna that parasitize cattle in each locality and farm. The difference can also be attributed to each farmer’s management of pasture livestock. In a given bioclimatic region, the latter factor is closely related to tick infestation of the cattle. These results were correlated with the presence of tick vectors of A. marginale and A. phagocytophilum, mainly Hyalomma spp., Rhipicephalus spp. and Ixodes spp. [46, 47].
A significant difference was observed in A. marginale infections between cattle breeds (P < 0.05), with the highest prevalence in the Schwytz breed. In Uganda, Magona and Mayende [48] reported a high rate of mortality in Friesian cattle due to a high prevalence of A. marginale associated or not with other pathogens (Theileria and Trypanosoma). Our results contradict those reported in Morocco by Ait Hamou et al. [45], where they described a non-significant difference in the prevalence of infection with A. marginale among cross, local and imported breeds. In fact, in North Africa, local and cross-breed animals are considered more resistant to anaplasmosis than pure breeds imported from Europe. But it seems that this relative resistance is due less to the breed than to the fact that animals born in endemic areas acquire a natural immunity (premunition) at an early age [44].
The age of cattle appears to influence the prevalence of anaplasmosis, A. marginale infection rate being significantly higher in older animals. Similarly, in Morocco, Ait Hamou et al. [45] reported the difference in the prevalence of A. marginale infections in calves (26.1 %) and adults (52.4 %). Our results were consistent with those reported in Uganda [48]. This difference might be explained by the more sustained exposure of adults to tick vectors [49]. Moreover, it appears that calves are less susceptible to the disease. Indeed, anaplasmosis is rare in animals younger than six months, while those between six months and one year usually develop only a mild illness, and cattle between one and two years old develop multiple signs of the disease, which is rarely fatal. However, the disease is often fatal after acute infection in adults over two years old, with a mortality risk ranging from 29 % to 49 % [7, 50]. Calves are temporarily protected (maternal antibodies) by the colostrum and a mother’s immunity, preventing short-term protection.
In our study, the prevalence of A. phagocytophilum (0.6 %) was much lower than that of A. marginale. This concurred with the results reported in Turkey where the prevalence of A. marginale (2.8 %) was higher than that of A. phagocytophilum (1 %) [41] but contradicted the study carried out in Italy by Torina et al. [51], where the prevalence of A. phagocytophilum (16.6 %) was higher than that of A. marginale (9.8 %). This difference may be attributed to (i) the fluctuation of the bacteraemia during the chronic phase of A. phagocytophilum and A. marginale infection [52]; (ii) the low number of intragranulocytic A. phagocytophilum circulating in carrier animals [53]; (iii) the fact that A. marginale develops faster than A. phagocytophilum in the host [54]; (iv) to the higher density of competent vectors and/or reservoirs; or (v) to the different tick infection rates by A. marginale and A. phagocytophilum.
Sequencing results showed a genetic variability in Tunisian A. marginale consistent with the great A. marginale genetic diversity found in endemic areas worldwide [18]. Presence of overlapped peaks at certain positions of four of the samples (sequences KR871279, KR871284, KR871281, KR871283) even after re-sequencing would suggest that certain animals harbored double infections, a circumstance already reported [55].
Conclusions
In conclusion, the duplex assay used here offered a rapid, specific and inexpensive mean of discriminating between A. marginale and A. phagocytophilum in carrier cattle. We now use this system routinely in our research and have tested it on blood samples, ticks, and fleas (data not shown). This technique could be used to detect these pathogens in tick vectors whose activity and abundance are affected by global warming [56]. The present study also determined the prevalence of the two pathogens and identified different A. marginale variants in cattle from different areas in Tunisia. More epidemiological studies are needed to determine the vectors and reservoir animals for the Anaplasma species and to clarify the pathogenicity of A. marginale and A. phagocytophilum for humans and animals in Tunisia.
Abbreviations
- A:
-
Anaplasma
- BLAST:
-
Basic local alignment search tool
- cELISA:
-
Competitive enzyme-linked immunosorbent assay
- E:
-
Ehrlichia
- groEL :
-
Heat-shock gene
- msp2:
-
Major surface protein 2 gene
- msp4:
-
Major surface protein 4 gene
- MSP5:
-
Major surface protein 5
- MSP5:
-
Major surface protein 5
- MSPs:
-
Major surface proteins
- NCBI:
-
National Center for Biotechnology Information
- OIE:
-
The Office International des Epizooties
References
Kocan KM, Blouin EF, Barbet AF. Anaplasmosis control. Past, present, and future. Ann N Y Acad Sci. 2000;916:501–9.
OIE 2008, World Organisation for Animal Health (OIE). Mannual of standards for diagnostic tests and vaccines for terrestirial animals, 6th editian, 2008, p. 599–10.
Rogers RJ, Shiels IA. Epidemiology and control of anaplasmosis in Australia. J S Afr Vet Assoc. 1979;50:363–6.
Kuttler KL. Clinical and hematologic comparison of Anaplasma marginale and Anaplasma centrale infections in cattle. Am J Vet Res. 1966;27:941–6.
Uilenberg U. Other ehrlichiosis of ruminants. In: Woldehiwet Z, Ristic M, editors. Rickettsial and chlamydial diseases of domestic animals. Oxford: Pergamon Press; 1993. p. 293–332.
Matsumoto K, Joncour G, Davoust B, Pitel PH, Chauzy A, Collin E, et al. Anaplasma phagocytophilum infection in cattle in France. Ann N Y Acad Sci. 2006;1078:491–4.
Kocan KM, de la Fuente J, Guglielmone AA, Melendez RD. Antigens and alternatives for control of Anaplasma marginale infection in cattle. Clin Microbiol Rev. 2003;16:698–712.
Kuttler KL. Anaplasma infections in wild and domestic ruminants: a review. J Wildl Dis. 1984;20:12–20.
Bakken JS, Dumler JS, Chen SM, Eckman MR, Van Etta LL, Walker DH. Human granulocytic ehrlichiosis in the upper Midwest United States. A new species emerging? JAMA. 1994;272:212–8.
Woldehiwet Z, Scott GR. Stages in the development of Cytoecetes phagocytophila, the causative agent of tick-born fever. J Comp Path. 1982;92:469–74.
Kocan KM, de la Fuente J, Blouin EF, Coetzee JF, Ewing SA. The natural history of Anaplasma marginale. Vet Parasitol. 2010;167:95–107.
Kocan KM, de la Fuente J, Blouin EF, Garcia-Garcia JC. Anaplasma marginale (Rickettsiales: Anaplasmataceae): recent advances in defining host-pathogen adaptations of a tick-borne rickettsia. Parasitol. 2004;129:S285–300.
Luther DG, Cox HU, Nelson WO. Comparison of serotests with calf inoculations for detection of carriers in anaplasmosis-vaccinated cattle. Am J Vet Res. 1980;41:2085–6.
Knowles D, Torioni de Echaide S, Palmer G, McGuire T, Stiller D, McElwain T. Antibody against an Anaplasma marginale MSP5 epitope common to tick and erythrocyte stages identifies persistently infected cattle. J Clin Microbiol. 1996;34:2225–30.
Dreher UM, de la Fuente J, Hofmann-Lehmann R, Meli ML, Pusterla N, Kocan KM, et al. Serologic cross-reactivity between Anaplasma marginale and Anaplasma phagocytophilum. Clin Diagn Lab Immunol. 2005;12:1177–83.
Strik NI, Alleman AR, Barbet AF, Sorenson HL, Wamsley HL, Gaschen FP, et al. Characterization of Anaplasma phagocytophilum major surface protein 5 and the extent of its cross-reactivity with A. marginale. Clin Vaccine Immunol. 2007;14:262–8.
Palmer GH, Barbet AF, Cantor GH, McGuire TC. Immunization of cattle with the MSP-1 surface protein complex induces protection against a structurally variant Anaplasma marginale isolate. Infect Immun. 1989;57:3666–9.
de la Fuente J, Ruybal P, Mtshali MS, Naranjo V, Shuqing L, Mangold AJ, et al. Analysis of world strains of Anaplasma marginale using major surface protein 1a repeat sequences. Vet Microbiol. 2007;119:382–90.
Park HS, Lee JH, Jeong EJ, Park TK, Kim TY, Chae JS, Park JH, Klein TA, Jang WJ, Park KH, Lee SH, et al. Differentiation of Anaplasmataceae through partial groEL gene analysis. Microbiol Immunol. 2005;49:655–62.
Dahmani M, Davoust B, Benterki MS, Fenollar F, Raoult D, Mediannikov O. Development of a new PCR-based assay to detect Anaplasmataceae and the first report of Anaplasma phagocytophilum and Anaplasma platys in cattle from Algeria. Comp Immunol Microbiol Infect Dis. 2015;39:39–45.
Reinbold JB, Coetzee JF, Sirigireddy KR, Ganta RR. Detection of Anaplasma marginale and A. phagocytophilum in bovine peripheral blood samples by duplex real-time reverse transcriptase PCR assay. J Clin Microbiol. 2010;48:2424–32.
Hurtado A, Barandika JF, Oporto B, Minguijón E, Povedano I, García-Pérez AL. Risks of suffering tick-borne diseases in sheep translocated to a tick infested area: a laboratory approach for the investigation of an outbreak. Ticks Tick Borne Dis. 2015;6:31–7.
Sarih M, M’ghirbi Y, Bouattour A, Gern L, Baranton G, Postic D. Detection and identification of Ehrlichia spp. in ticks collected in Tunisia and Morocco. J Clin Microbiol. 2005;43:1127–32.
M’ghirbi Y, Ghorbel A, Amouri M, Nebaoui A, Haddad S, Bouattour A. Clinical, serological, and molecular evidence of ehrlichiosis and anaplasmosis in dogs in Tunisia. Parasitol Res. 2009;104:767–74.
M’Ghirbi Y, Yaïch H, Ghorbel A, Bouattour A. Anaplasma phagocytophilum in horses and ticks in Tunisia. Parasit Vectors. 2012;5:180–4.
Belkahia H, Ben Said M, Sayahi L, Alberti A, Messadi L. Detection of novel strains genetically related to Anaplasma platys in Tunisian one-humped camels (Camelus dromedarius). J Infect Dev Ctries. 2015;9:1117–25.
Belkahia H, Ben Said M, Alberti A, Abdi K, Issaoui Z, Hattab D, et al. First molecular survey and novel genetic variants' identification of Anaplasma marginale, A. centrale and A. bovis in cattle from Tunisia. Infect Genet Evol. 2015;34:361–71.
Ben Said M, Belkahia H, Alberti A, Zobba R, Bousrih M, Yahiaoui M, et al. Molecular survey of Anaplasma species in small ruminants reveals the presence of novel strains closely related to A. phagocytophilum in Tunisia. Vector Borne Zoonotic Dis. 2015;15:580–90.
Ben Said M, Belkahia H, Karaoud M, Bousrih M, Yahiaoui M, Daaloul-Jedidi M, Messadi L. First molecular survey of Anaplasma bovis in small ruminants from Tunisia. Vet Microbiol. 2015;179:322–6.
Zeidner NS, Burkot TR, Massing R, Nicholson WL, Dolan MC, Rutherford JS, et al. Transmission of the agent of human granulocytic ehrlichiosis by Ixodes spinipalpis ticks: evidence of an enzootic cycle of dual infection with Borrelia burgdorferi in Northern Colorado. J Infect Dis. 2000;182:616–9.
Rikihisa Y. New findings on members of the family Anaplasmataceae of veterinary importance. Ann NY Acad Sci. 2006;1078:438–45.
Ismail N, Bloch KC, McBride JW. Human ehrlichiosis and anaplasmosis. Clin Lab Med. 2010;30:261–92.
Doudier B, Olano J, Parola P, Brouqui P. Factors contributing to emergence of Ehrlichia and Anaplasma spp. as human pathogens. Vet Parasitol. 2010;167:149–54.
Singh H, Jyoti Haque M, Singh NK, Rath SS. Molecular detection of Anaplasma marginale infection in carrier cattle. Ticks Tick Borne Dis. 2012;3:55–8.
Sharma A, Singla LD, Tuli A, Kaur P, Bal MS. Detection and assessment of risk factors associated with natural concurrent infection of Trypanosoma evansi and Anaplasma marginale in dairy animals by duplex PCR in eastern Punjab. Trop Anim Health Prod. 2015;47:251–7.
De la Fuente J, Torina A, Caracappa S, Tumeno G, Furla R, Almazan C, Kocan KM. Serologic and molecular characterization of Anaplasma species infection in farm animals and ticks from Sicily. Vet Parasitol. 2005;133:357–62.
Pohl AE, Cabezas-Cruz A, Flávio M, Ribeiro B, Angélica J, da Silveira G, et al. Detection of genetic diversity of Anaplasma marginale isolates in Minas Gerais, Brazil. Rev Bras Parasitol Vet. 2013;22:129–35.
Mutshembele AM, Cabezas-Cruzc A, Mtshali MS, Thekisoe OMM, Galindo RC, de la Fuente J. Epidemiology and evolution of the genetic variability of Anaplasma marginale in South Africa. Ticks Tick borne Dis. 2014;5:624–31.
Hairgrove T, Schroeder ME, Budke CM, Rodgersb S, Chungd C, Uetie MW, Bounpheng MA. Molecular and serological in-herd prevalence of Anaplasma marginale infection in Texas cattle Preventive. Vet Med. 2015;119:1–9.
Shebish E, Vemulapalli R, Oseto C. Prevalence and molecular detection of Anaplasma marginale, Babesia bovis and Babesia bigemina in cattle from Puntarenas Province, Costa Rica. Vet Parasitol. 2012;188:164–7.
Aktas M, Altay K, Dumanli N. Molecular detection and identification of Anaplasma and Ehrlichia species in cattle from Turkey. Ticks Tick Borne Dis. 2011;2:62–5.
El-Ashker M, Hotzelb H, Gwidab M, El-Beskawyd M, Silaghi C, Tomaso H. Molecular biological identification of Babesia, Theileria, and Anaplasma species in cattle in Egypt using PCR assays, gene sequence analysis and a novel DNA microarray. Vet Parasitol. 2015;207:329–34.
Ybanez AP, Ybanez RHD, Cruz-Flores MJ, Xuenan X, Yokoyama N, Inokuma H. High genetic diversity of Anaplasma marginale detected from Philippine cattle. J Vet Med Sci. 2014;76:1009–14.
Sergent E, Donatien A, Parrot L, Lestoquard F. Etude sur les Piroplasmoses Bovines. Arch Inst Pasteur Algérie. 1945. p. 816.
Ait Hamou S, Rahali T, Sahibi H, Belghyti D, Losson B, Goff W, Rhalem A. Molecular and serological prevalence of Anaplasma marginale in cattle of North Central Morocco. Res Vet Sc. 2012;93:1318–23.
Bouattour A, Darghouth MA, Daoud A. Distribution and ecology of ticks (Acari: Ixodidae) infesting livestock in Tunisia: an overview of eight years field collection. Parasitologia. 1999;41:5–10.
M’ghirbi Y, Hurtado A, Bouattour A. Theileria and Babesia parasites in ticks in Tunisia. Transboundary Emerging Dis. 2010;57:49–51.
Magona JW, Mayende JSP. Occurrence of concurrent trypanosomosis, theileriosis, anaplasmosis and helminthosis in Friesian, Zebu and Sahiwal cattle in Uganda. J Vet Res. 2002;69:133–40.
Bouattour A, Darghouth MA, Ben ML. Cattle infestation by Hyalomma ticks and prevalence of Theileria in Hyalomma. detritum species in Tunisia. Vet Par. 1996;65:256–63.
Richey EJ. Bovine anaplasmosis. In: The 24th Annual Convention of Proceedings of American Association of Bovine Practices, Orlando, Florida, 1991. p. 3–11.
Torina A, Vicente J, Alongi A, Scimeca A, Turla R, Nicosia S, et al. Observed prevalence of tick-borne pathogens in domestic animals in Sicily, Italy during 2003–2005. Zoon Pub Health. 2007;54:8–15.
Kieser ST, Eriks IE, Palmer GH. Cyclic rickettsemia during persistent Anaplasma marginale infection in cattle. Infect Immun. 1990;58:1117–9.
Pusterla N, Braun U. Clinical findings in cows after experimental infection with Ehrlichia phagocytophila. Zentralbl Veterinarmed. 1997;44:385–90.
Hoar BR, Nieto NC, Rhodes DM, Foley JE. Evaluation of sequential coinfection with Anaplasma phagocytophilum and Anaplasma marginale in cattle. Am J Vet Res. 2008;69:1171–8.
Palmer GH, Rurangirwa FR, McElwain TF. Strain composition of the ehrlichia Anaplasma marginale within persistently infected cattle, a mammalian reservoir for tick transmission. J Clin Microbiol. 2001;39:631–5.
Estrada-Pena A. Tick-borne pathogens, transmission rates and climate change. Front Biosci (Landmark Ed). 2009;14:2674–87.
Acknowledgements
We are grateful to Mr Adel Rhim, Dr. Leila Saieh, Dr. Kamel Khlif, Dr. Zouheir Ktata and Dr Ridha Ben Omrane for their help in field work. The authors would also thank all farmers for their collaboration in sample collections. A particular thank to Deborah Glassman for her constructive comments and English corrections of the manuscript.
Funding
This work was conducted with financial support from the Spanish Agency for International Development Cooperation (AECID, Project No. A/026818/09), the Spanish National Institute for Agricultural and Food Research and Technology (INIA, Project No. RTA2009-000-18-00-00), the European Regional Development Fund (ERDF), the IEVP coopération transfrontalière Italie-Tunisie 2007-2013 PROJET 2 PS1.3.023-RESTUS and the Tunisian Ministry for Higher Education, Scientific Research, and Technology.
Availability of data and materials
Sequence data were deposited in GenBank; accession numbers for the partial msp2 and msp4 sequences are KR871275–KR871287.
Authors’ contributions
YM analyzed the samples, interpreted data, wrote the paper and conducted the field survey. MB conducted the laboratory examination of samples and sequences analysis. BO was involved in the setting up of the duplex PCR. FK was involved in sequence analysis and submission. AH and AB revised the manuscript, designed and supervised the study. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The study has been approved by the Commission on Ethics and Animal Welfare of the Institut Pasteur de Tunis - Université Tunis El Manar-TUNISIA with the given number 2014/03/I/LR11IPT03/V1. All technical procedures were in accordance with the National and the European legislation regarding animal welfare and have met the International Guiding Principles for Biomedical Research Involving Animals by the Council for the International Organizations of Medical Sciences.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
M’ghirbi, Y., Bèji, M., Oporto, B. et al. Anaplasma marginale and A. phagocytophilum in cattle in Tunisia. Parasites Vectors 9, 556 (2016). https://doi.org/10.1186/s13071-016-1840-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-016-1840-7